Contents
IntroductionHints on Customising Your Picture
Ligand binding site
Nucleic acid
Annotation: Labelling the Structure
Script File Format
 |
CCP4 Molecular Graphics Documentation | |
| Picture Wizard |
| Documentation Contents | On-line Documentation | Tutorials | CCP4mg Home |
The Picture Wizard is a tool to quickly set up complex pictures. The Picture Wizard interface shows various representations of molecular models - you can select a representation style and it will be applied to your loaded model. New display objects will be created for the model in the same style as those shown in the interface pictures.
The representation styles are organised into Style folders such as 'nucleic acids' and 'ligand binding site'. The data files for the different representations are in the directory ccp4mg/data/picture_wizard_files.
There is an interface to the Picture Wizard at the top of the file browser for Open data file.. and it can be accessed from the icon menu of a model object. The Picture Wizard will analyse the data to identify ligands, nucleic acid etc. in order to set up a picture automatically. For a few styles, such as interfaces, you will be required to select one or more sets of atoms. If you click the Open Picture Wizard choices option on the file browser you will see this interface in which you can select a limited set of atoms for a scene.
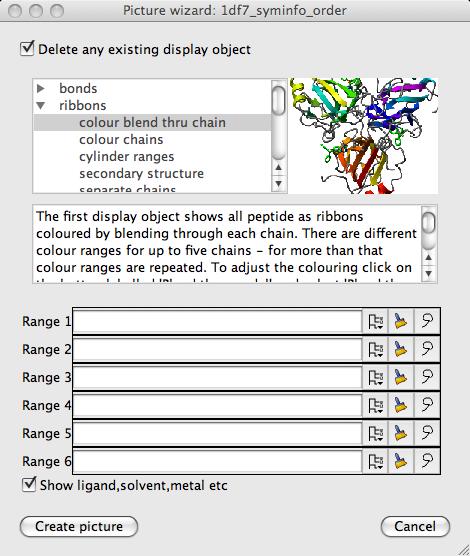
This window has some standard options:
Delete any existing display objects By default this is on, existing display objects are deleted when the new display objects for the picture are created.
The Picture Wizard is also accessible from the Application pull-down menu. In this version the chosen representation is aplied to all currently visbible models. Another difference is that the options to select specific sets of atoms (e.g. specific ligands) is not available. Some of the representation modes which would require user selection of sets of atoms (particularly for interfaces) are not available here.
After creating a picture with the Wizard you may want to change some aspect of the picture. It is generally easy to change the colour and display style of display objects so the representations available in the Wizard focus on examples of different atom selections and leave you to set the colour and display style.
You may want to hide or delete some display object; use the options display object icon menu ![]() to do this. In the Create picture window is the option to Keep current display objects; you could use this to set up display objects from more that one picture wizard representation; for example if your 'ligand' is nucleic acid you may want to use a ligand binding site representation and a nucleic acid representation.
to do this. In the Create picture window is the option to Keep current display objects; you could use this to set up display objects from more that one picture wizard representation; for example if your 'ligand' is nucleic acid you may want to use a ligand binding site representation and a nucleic acid representation.
Selecting the appropriate residues to display for an binding site is difficult to automate completely; it requires a human to know which residues are important biologically and to judge whether a picture is 'over complex'. The wizard ligand binding site pictures use up to three display objects with neighbourhood selections to define a binding site:
A selection of main chain/side chain close to the ligand (usually with at least one atom within 3 Å of the ligand). Note that using this mode rather than residue mode is usually effective in removing some main chain where only the side chain interacts with the ligand and vica versa.
A selection of main chain/side chain with hydrogen bonding capability to the ligand. The CCP4mg default cutoff for display of hydrogen bonds is usually 3.9Å which is very generous. To ensure no potential hydrogen bonding groups are lost the neighbourhood selection cutoff is usually 4.0Å.
To give some idea of the context of the binding site the whole structure can be displayed as ribbons or short stretches of main chain around the site can be shown (displayed as worms seems appropriate). A display object selecting neighbourhood main chain within 6-7Å of the ligand is used in some Wizard pictures.
If the automatic selection does not give the result you require you can try tweaking the cutoff distances but will probably have to use the Interactive Selection option to add or remove individual atoms or residues.
If you create a picture with a surface the surface will initailly be hidden - you need to click on the surface icon ![]() with the right mouse button to make it visible. Calculating the surface may take some time. All surfaces will initially be drawn opaque; to make a surface transparent select Set transparency from the View menu and set an opacity level for the surface object (about 0.3 is usually good) and then toggle on the Transparency on/off checkbox. Beware that drawing transparent surfaces is very slow and you are best to toggle transparancy off before changing the opacity level, changing the view or doing anything else.
with the right mouse button to make it visible. Calculating the surface may take some time. All surfaces will initially be drawn opaque; to make a surface transparent select Set transparency from the View menu and set an opacity level for the surface object (about 0.3 is usually good) and then toggle on the Transparency on/off checkbox. Beware that drawing transparent surfaces is very slow and you are best to toggle transparancy off before changing the opacity level, changing the view or doing anything else.
Most nucleic acid representations use 'base blocks' to fill in the bases with flat blocks. The base blocks are usually displayed in conjunction with a cylinders representation of the structure. Colouring the blocks different from the cylinders can be effective. The cylinders could also be changed to ball-n-stick though the stick thickness will probably need increasing. The backbone of the nucleic acid can be drawn as ribbon and in this case it often looks better if any accompanying cylinders exclude the backbone from the selection to minimise the bits of cylinder 'sticking out' of the ribbon.
Some Wizard pictures have the option to label binding site residues. These labels are in a fixed position relative to the residue Ca atom and have a limited range of formats. The labels can be toggled on or off, or the displayed information changed from the display object icon menu ![]() options Label atoms and Label text. The font used for these labels cn be changed: in the Preferences window, in the Display folder, select Fonts and change the Atom label font.
options Label atoms and Label text. The font used for these labels cn be changed: in the Preferences window, in the Display folder, select Fonts and change the Atom label font.
An alternative approach to labelling the structure which gives more flexiblity in content and position of the text is Annotation.
The user-created picture wizard script files are called .CCP4MG/data/picture_wizard_files/style_folder/style_name.mgpic.py. The general format of the file has three sections:
#SECTION TITLE The title that appears under the snapshot in the interface (keep it short!) #SECTION NOTES The notes that the user can access from the interface #SECTION CHOICES A list of the options that will appear in the Create picture window (e.g. selecting data objects, ligands, chains). #SECTION SCRIPT A Python language script to create the picture
The options which appear in the Create picture interface are specified in the CHOICES section of the script file. This section specifies a list of variables which all have, at least, a name, a type and a label. The type and the label information are used to create an appropriate interface. A variable with the given name is initialised with the user's chosen value before the script is run so the script can 'see' the user's choices.
The SCRIPT section is a picture definition file with the display object attributes. The picture definition file is usually used to save the complete program status and includes definition of the data objects (particularly the name of the file that data should be loaded from). The picture wizard files do not have any definition of data objects as these are expected to be already loaded into CCP4mg. The picture wizard file defines the display objects - for example a MolDisp (model display object):
MolDisp ( parent = MolData1,
colour = 'atomtype',
style = 'BALLSTICK',
selection_parameters = {
'select' : 'nopeptide',
'monomers' : monomers_list } )
This is a command to create a MolDisp (model display) object; the command has several arguments to define the object. The most important argument is parent, the name of its parent data object. The value for parent is in the variable MolData1; the value of this variable is initialised before the script is run from the users choices. MolData1 is one of the variables which must be defined in the CHOICES section. The other arguments determine the atom selection, colour and display style for the object and are explained in the object attributes documentation.
An example of a CHOICES section
CHOICES = [
{ 'name' : 'MolData1','type' : 'MolData','label' : 'Select model','multi' : 0 },
{ 'name' : 'ligand1', 'type' : 'monomer','label' : 'Ligand 1', 'parent':'MolData1' },
{ 'name' : 'ligand2', 'type' : 'monomer','label' : 'Ligand 2', 'parent':'MolData1' },
{ 'name' : 'label_site_residues', 'type' : 'logical','label' : 'Label binding site residues' , 'initial' : '0' }
]
This section is in Python syntax and defines a list called CHOICES. The items in the list are dictionaries. See the Python syntax page for an explanation of the Python syntax for lists and dictionaries.
An auto-generated script file created if you Save current representation includes the option to select the data object(s) in the CHOICES section but will probably include some commands in the SCRIPT section which are specific to the data objects that were used creating the files, for example the specific names of ligands or residue ranges. These specific selections probably will need replacing by variables that are specified in the CHOICES section.