Stereo Views of Figures 1-5 from "An Overview of ProSMART"
(return to article)To view image in full size, right-click on the image and select open/view/save image, depending on the browser.
(ii) 3b50_A
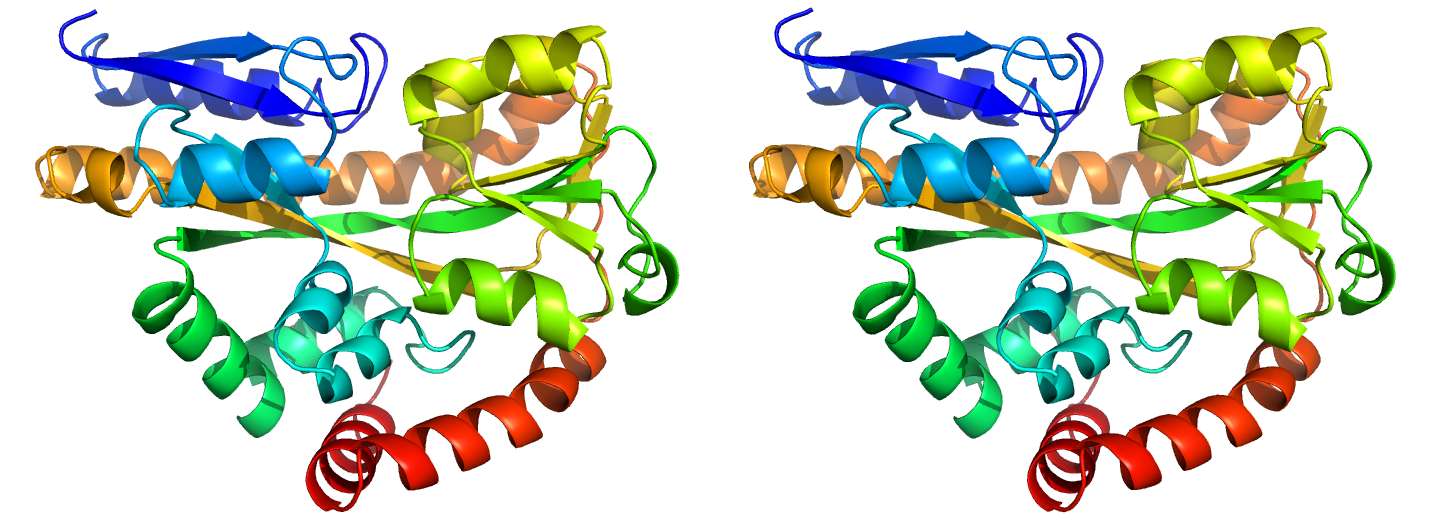
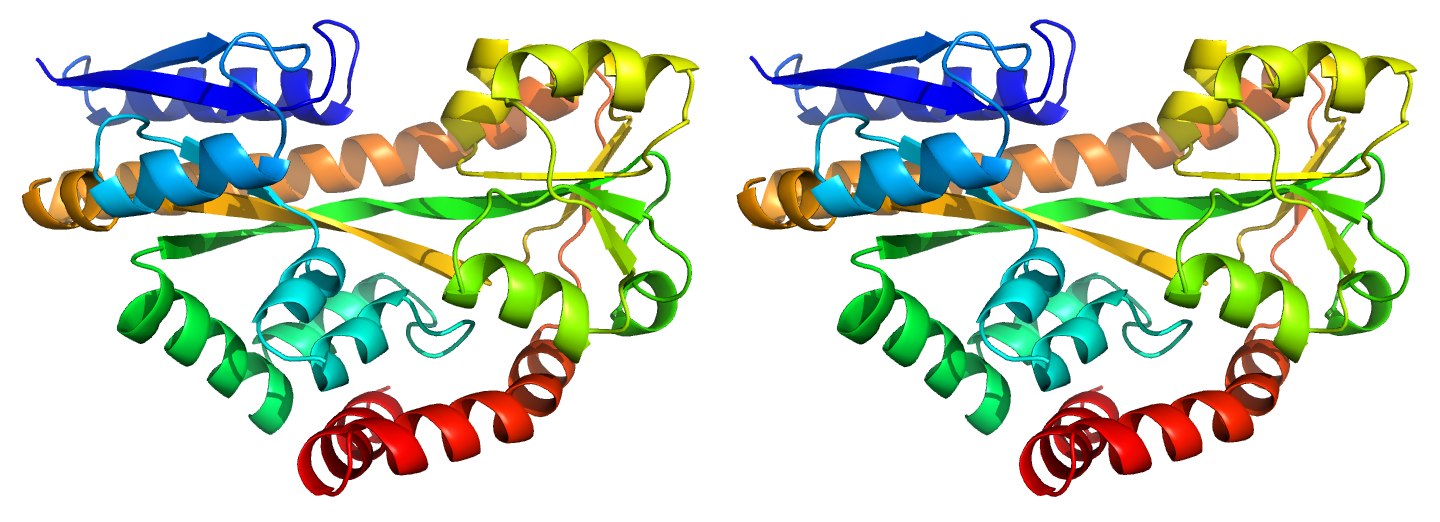
Figure 1. Illustrations of sequence-identical chains, specifically the open (i, 2cex_A) and closed (ii, 3b50_A) forms of the SiaP TRAP SBP (Fischer et al., 2010), rainbow-coloured along the chain.
(ii)
(iii)
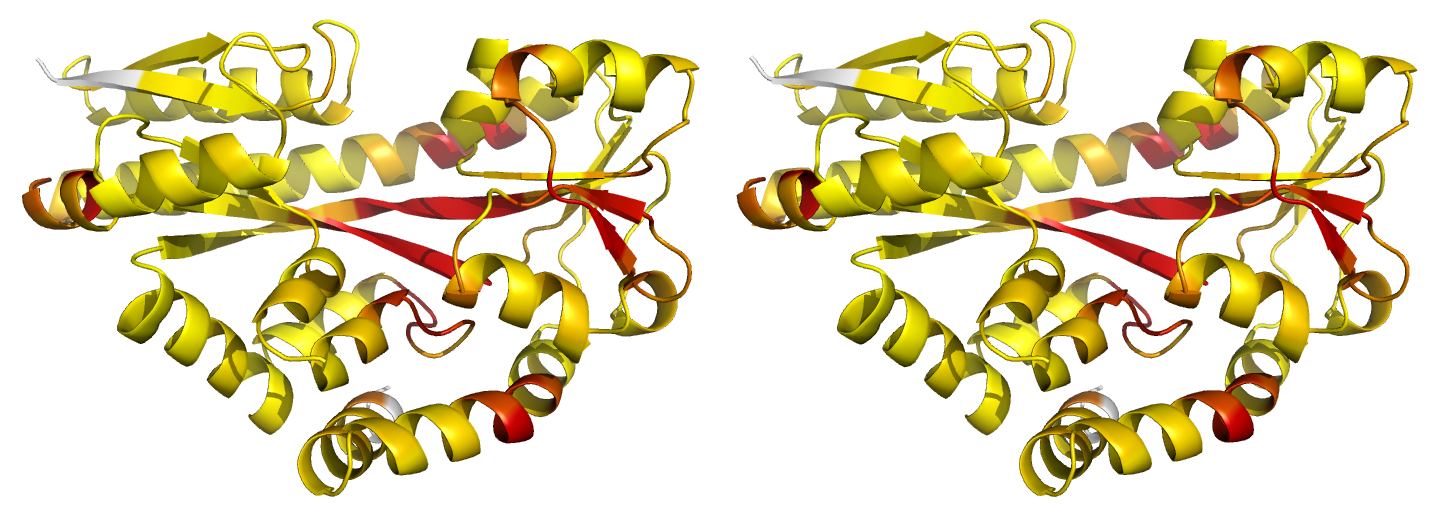
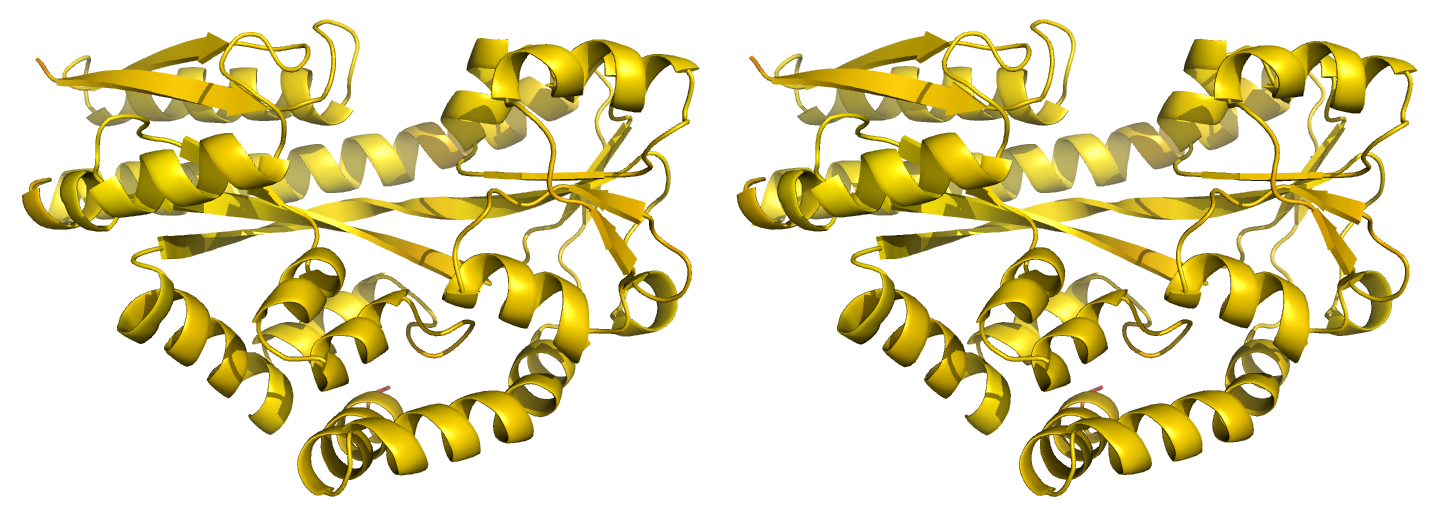
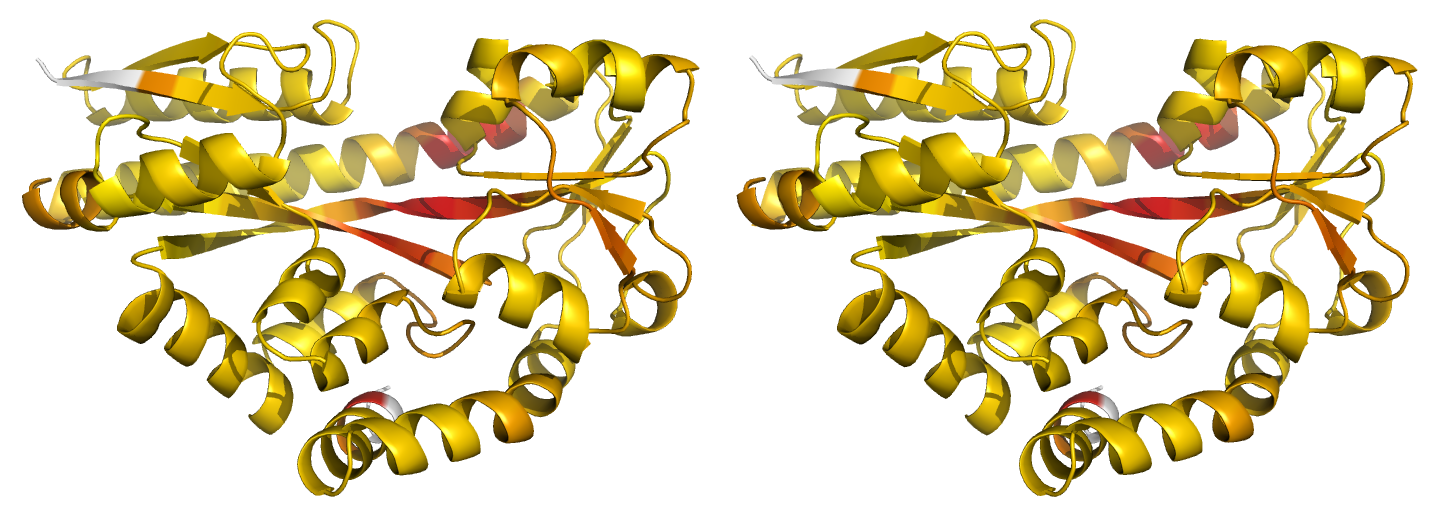
Figure 2. Illustrations of simple results from the default ProSMART comparison of 2cex_A and 3b50_A, coloured using a gradual colour gradient according to main-chain dissimilarity scores (yellow implies similarity, red relative dissimilarity). These scores allow quick visual identification of exactly which regions are structurally very similar, and which exhibit differences. The "minimum score" (i) is highly insensitive to global conformation - note that all residues are aligned and identified as very similar despite the global conformational change. The "central score" (ii) is more sensitive to differences in local structural environment - note that locally distorted regions such as the hinge are easily identified. The "intrafragment rotational dissimilarity score" (iii) is sensitive to curvature and torsion of the local backbone - this score is useful for identifying regions that exhibit subtle backbone deformations that can be very hard to otherwise identify or quantify.
(ii)
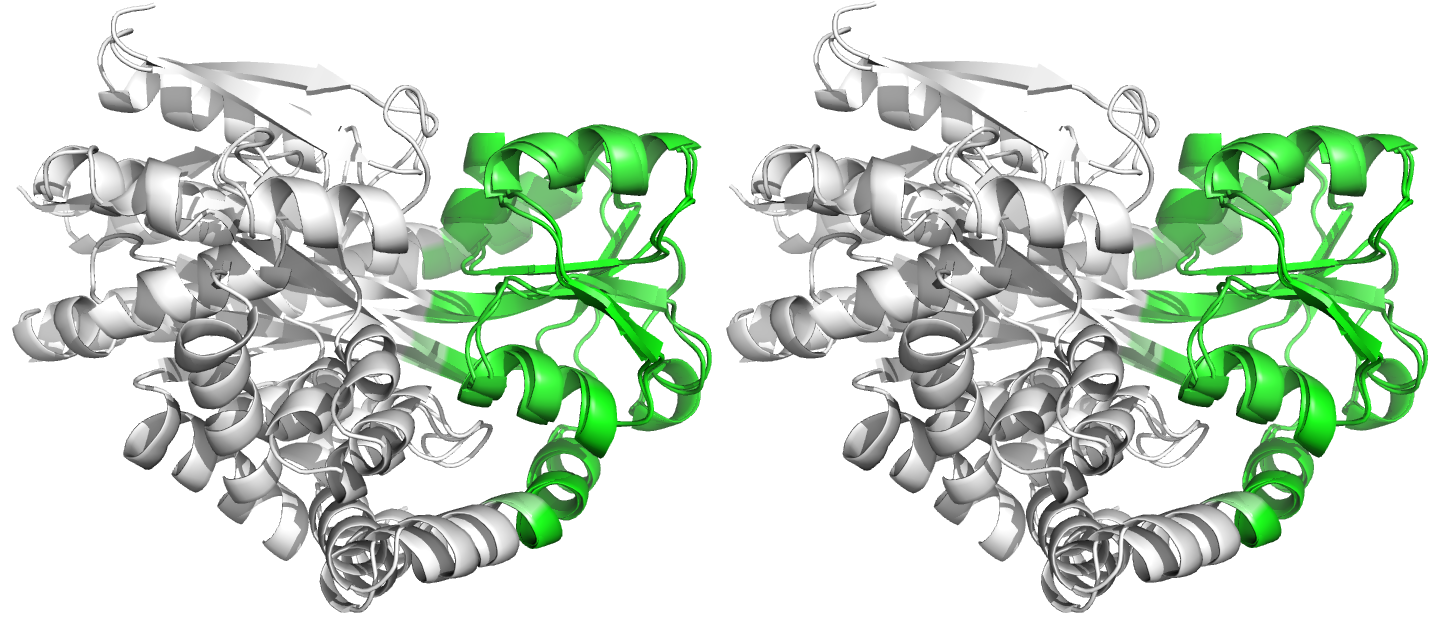
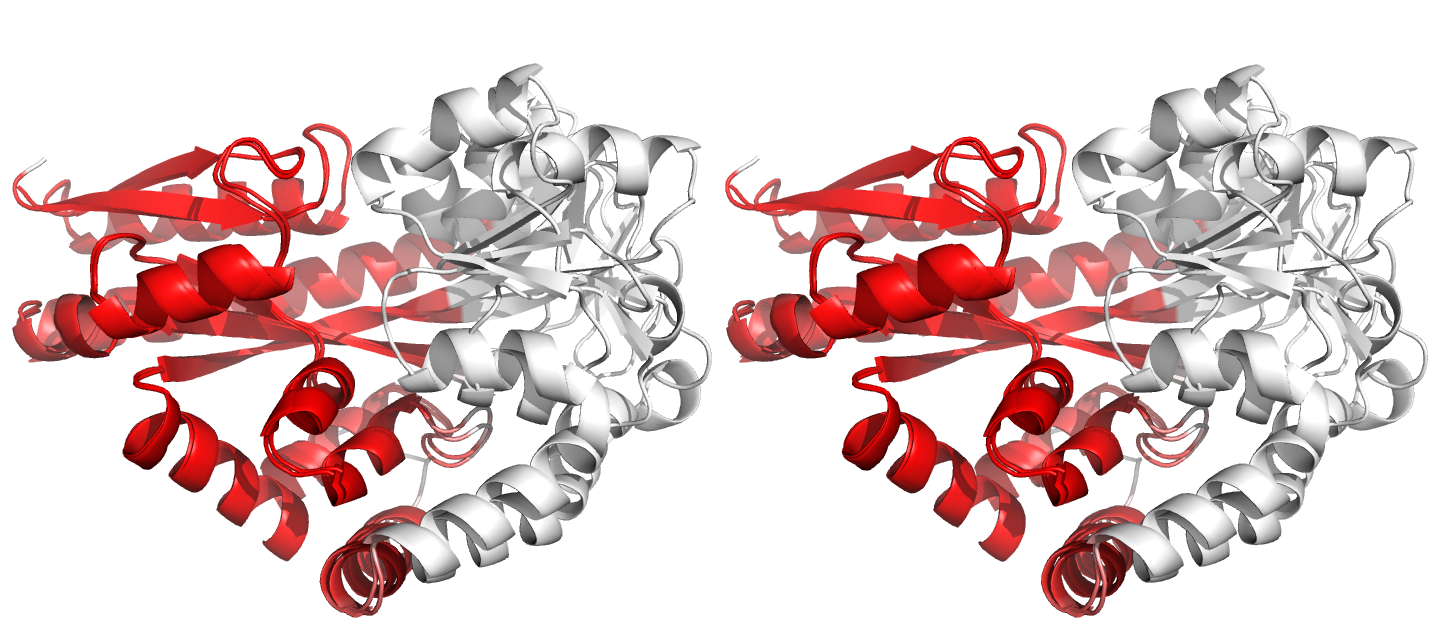
Figure 3. Superpositions arising from the rigid substructure identification results from default ProSMART comparison of 2cex_A and 3b50_A, coloured according to cluster scores. Two rigid substructures identified, coloured red (i) and green (ii), corresponding to the two domains.
(ii)
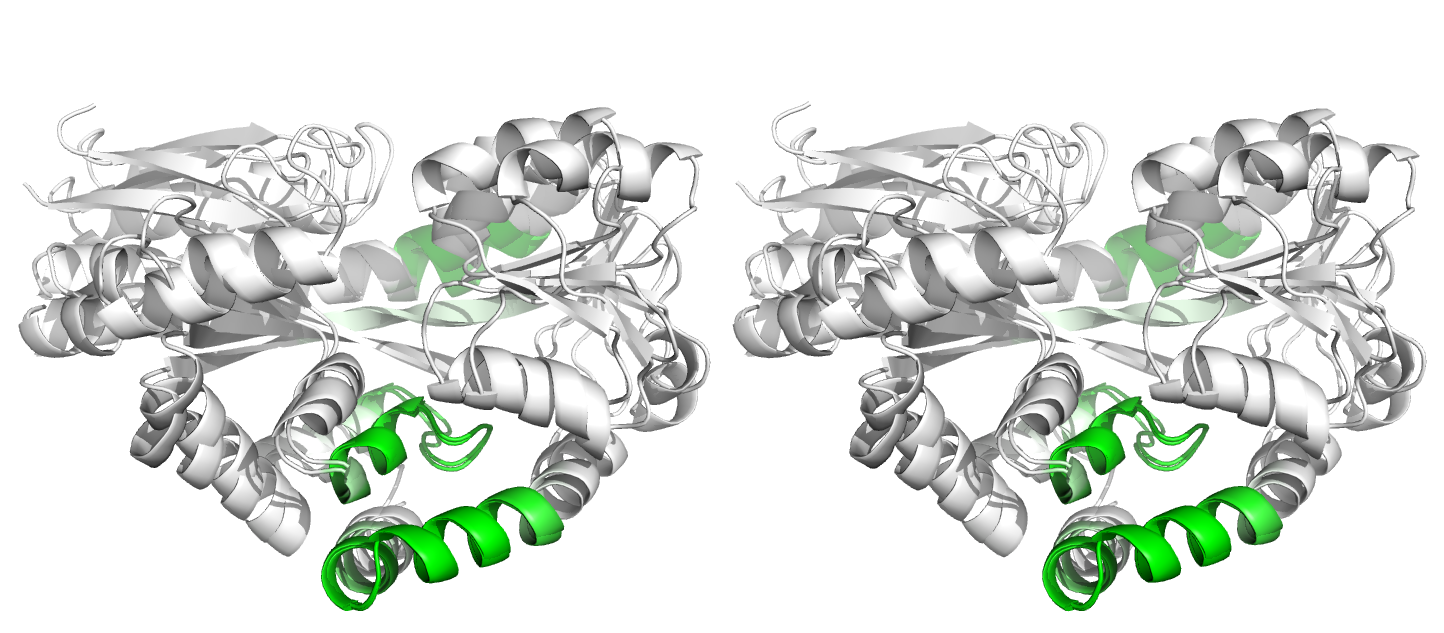
(iii)
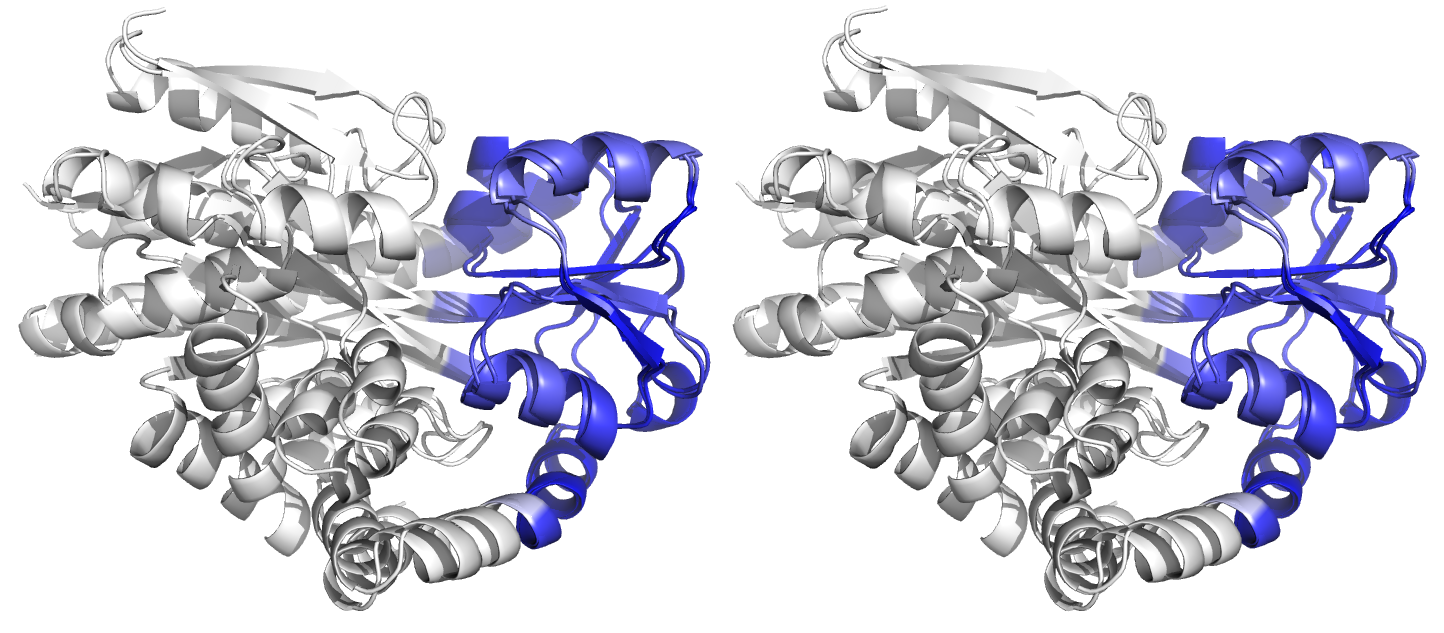
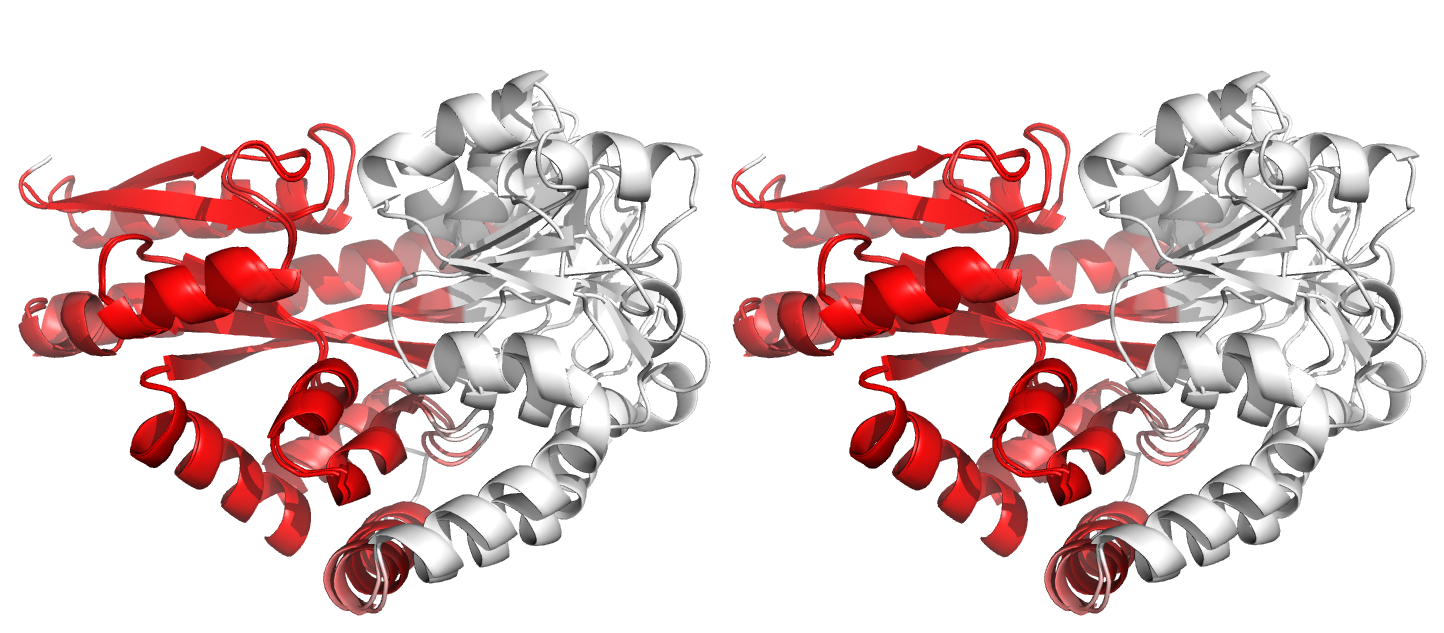
Figure 4. Superpositions arising from the rigid substructure identification results from the ProSMART comparison of 2cex_A and 3b50_A using a fragment length of 7 residues (keyword: -len), coloured according to cluster scores. In contrast with Figure 3, which used the default fragment length of 9 residues, three rigid substructures are identified, coloured red (i), green (ii) and blue (iii). These substructures correspond to two domains and the hinge region. This helps to illustrate the utility of performing comparative analyses at multiple levels of structural resolution.
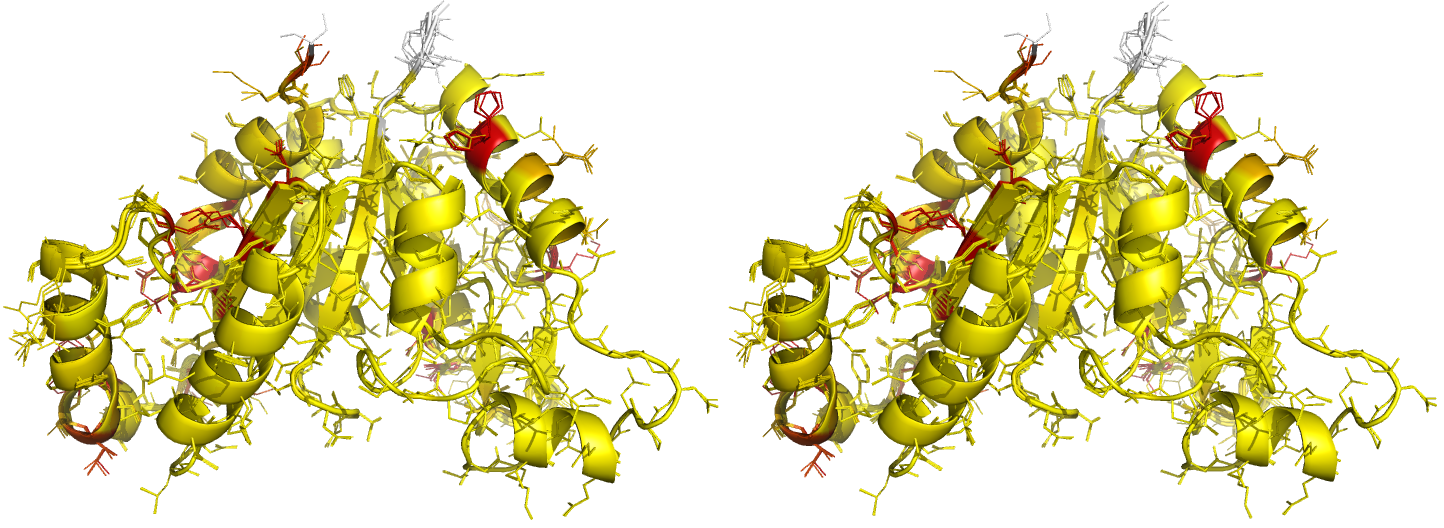
Figure 5. Illustrations of superposed NCS-related chains in the structure of BioD with PDB code 3MLE (Porebski et al., 2012). Residues are coloured according to side-chain RMSD relative to the local coordinate frame, allowing easy visual identification of residues with side-chains in different conformations. This can be particularly useful for cross-validation during various stages of the refinement process (e.g. by identifying changes in side-chain conformation before/after refinement, and identifying which side-chains are/aren't pulled towards reference structures after application of external side-chain restraints).
(return to article)