An Overview of ProSMART
, Marcus Fischer and Garib N. Murshudov
MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 0QH
Email: nicholls@mrc-lmb.cam.ac.uk
"Procrustes owned two
beds, one small, one large; he made short victims lie in the large bed, and the
tall victims in the short one…" (Taleb, 2010)
Procrustes was a mythological Greek villain whose victims were stretched and cut in
order to fit the shape of his bed. The "Pro" in ProSMART is due to its use of Procrustes
analysis (Gower, 2010; Gower and Dijksterhuis, 2004; Catell and Hurley, 1962) for comparing local regions of
structure between two protein chains. The name is fitting in this context due
to manipulating the coordinates from one structure in order to optimally fit
those in another, efficiently achieving a measure of local main-chain r.m.s.d. at the chosen level of structural resolution. By
performing an exhaustive structural comparison of all local regions between two
input structures, ProSMART is able to produce
alignments by optimising the net agreement of local
structures along the chain. The resulting alignment is thus independent of the
global conformation of the compared chains - this is subsequently exploited for
various purposes…
Contents
- Introduction
- Structural
Comparison Features
- External
Restraints for use in Macromolecular Crystallographic Refinement
- Choice of external reference
structure(s)
- Selection of suitable
structural information
- Important
REFMAC5 parameters
- Generic fragment and
secondary-structure-based restraints
- How to Run ProSMART
- Command
line
- CCP4i
REFMAC5 GUI
- How to ensure that the external
restraints are being used by REFMAC5 during refinement
- How
to use the REFMAC5 GUI with a pre-generated ProSMART
restraints file
- CCP4i
ProSMART GUI
- ProSMART Output
- Availability
and Contact Information
- Acknowledgements
- References
Introduction
ProSMART (Procrustes Structural Matching Alignment and Restraints
Tool) has two main purposes: conformation-independent comparison of protein
structures, and the generation of interatomic
distance restraints for subsequent use in macromolecular crystallographic
refinement by REFMAC5 (Murshudov et al., 2011, 1997; Nicholls et al.,
2012). Therefore, the tool comprises two major components:
- ProSMART ALIGN - for
conformation-independent structural comparison;
- ProSMART RESTRAIN - for external restraint
generation.
ProSMART is
written in C++, takes one or many PDB files as input, and can be run from the
command line or using a CCP4i (Potterton et al., 2003) interface (see below). Mac/Linux and Windows versions are available.
This article gives a brief overview of some of the
features available in ProSMART, a discussion
regarding the generation of external structural restraints, an overview of the
ways to run ProSMART, and the nature of the output.
Structural Comparison Features
Several features are available for performing different
types of comparative structural analyses, depending on the level of structural
similarity of the compared protein chains. ProSMART
is particularly well suited to the comparative analysis of homologous chains in
different global conformations, e.g. apo versus holo. Further to being able to achieve an alignment between
similar structures, you could in principle use ProSMART
to create an optimal alignment between completely dissimilar structures. The
alignment achieved by ProSMART is effectively the
optimal conformation-independent net agreement of local structures along the
chain; alignment filtering according to local structural dissimilarity scores
may be subsequently performed, if desired.
Further to achieving a conformation-independent
alignment, ProSMART automatically performs
identification and superposition of rigid substructures that are conserved
between the compared chains.
ProSMART uses
various residue-based scores for describing the dissimilarity of aligned
residues' local structural environments. These scores include measures that are
robust, allowing the identification of similarity in the presence of
conformational change. Other scores are very sensitive, being able to detect
subtle changes in the local structural environments that would otherwise be
extremely hard to detect. These scores complement each other, maximising the amount of information achieved when
performing such structural analyses (see Figure 2).
Another major feature of ProSMART
is that the default generated output allows publication-quality illustrations
to be quickly and easily achieved using the molecular graphics software CCP4mg
(McNicholas et al., 2011) or PyMOL
(Schrödinger; DeLano, 2002), for various types
of comparative structural analyses. For example, Figures 2-5 display default
results from ProSMART in PyMOL,
without any subsequent manipulation of superpositions
or colouring (the illustrations were not tediously
prepared by hand!).
The structural comparison methods used in ProSMART will be described in a future article (in the
meantime, details of the methods are described by Nicholls, 2011). For the
purposes of this article, we shall provide a few figures to briefly illustrate
some of the functionalities that may be applied to highly homologous
chain-pairs (thus would be of most relevance to cases involving external
restraint generation).
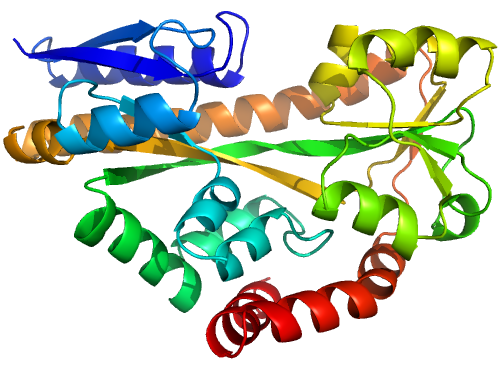
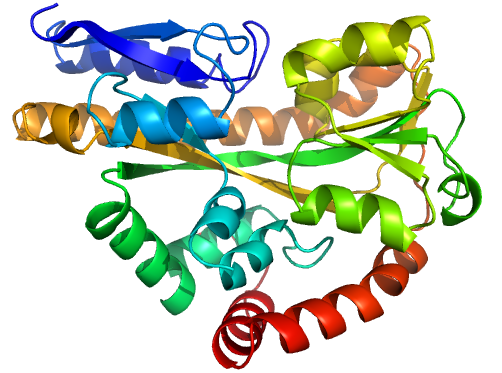
Figure 1 (stereo). Illustrations of sequence-identical
chains, specifically the open (2cex_A, left) and closed (3b50_A, right) forms
of the SiaP TRAP SBP (Fischer et al., 2010), rainbow-coloured along the chain.
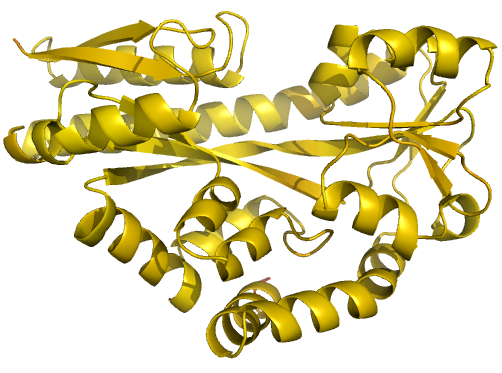
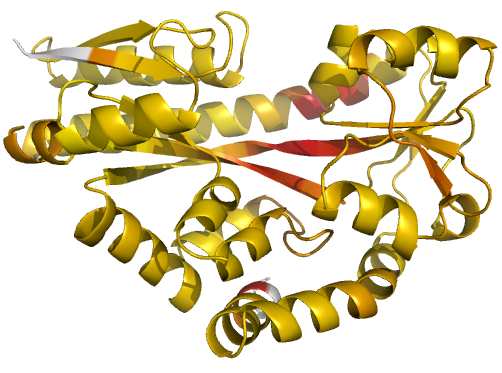
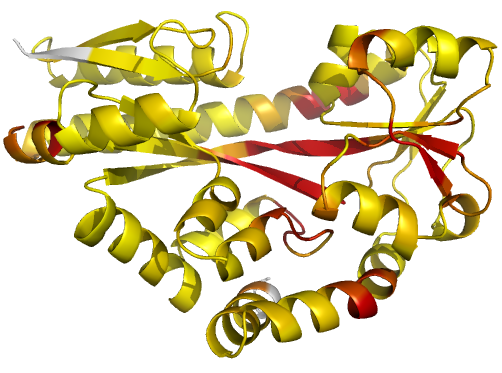
Figure 2 (stereo). Illustrations of simple
results from the default ProSMART comparison of
2cex_A and 3b50_A, coloured using a gradual colour gradient according to main-chain dissimilarity
scores (yellow implies similarity, red relative dissimilarity). For clarity,
only the chain 2cex_A is shown (if shown, residues in 3b50_A would have been coloured the same as the corresponding residues in 2cex_A).
These depictions allow quick visual identification of exactly which regions are
structurally very similar, and which exhibit differences. The "minimum
score" (left) is highly insensitive to global conformation - note that all
residues are aligned and identified as very similar despite the global
conformational change. The "central score" (middle) is more sensitive
to differences in local structural environment - note that locally distorted
regions such as the hinge are easily identified. The "intrafragment
rotational dissimilarity score" (right) is sensitive to curvature and
torsion of the local backbone - this score is useful for identifying regions
that exhibit subtle backbone deformations that can be very hard to otherwise
identify or quantify.
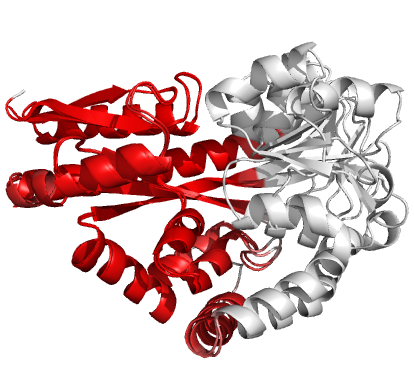

Figure 3 (stereo). Superpositions
arising from the rigid substructure identification results from default ProSMART comparison of 2cex_A and
3b50_A, coloured according to cluster scores. Two
rigid substructures identified, coloured red (left)
and green (right), corresponding to the two domains.
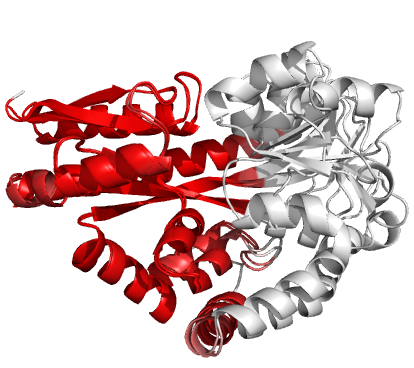
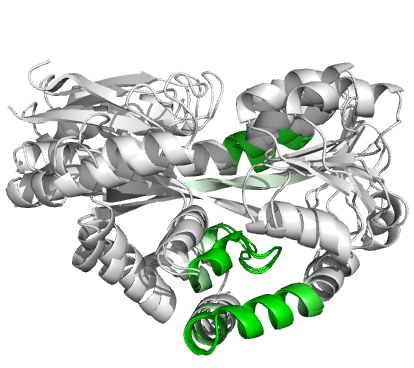
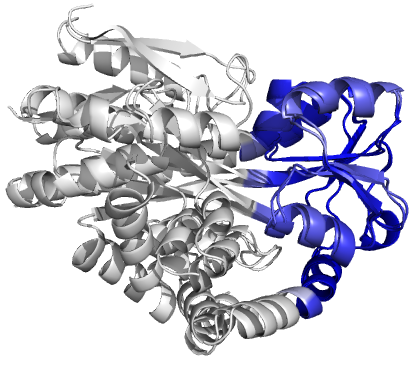
Figure 4 (stereo). Superpositions
arising from the rigid substructure identification results from the ProSMART comparison of 2cex_A and 3b50_A using a fragment
length of 7 residues (keyword: -len), coloured according to cluster scores. In
contrast with Figure 3, which used the default fragment length of 9 residues,
three rigid substructures are identified, coloured
red (left), green (middle) and blue (right). These substructures correspond to
two domains and the hinge region. This helps to illustrate the utility of
performing comparative analyses at multiple levels of structural resolution.
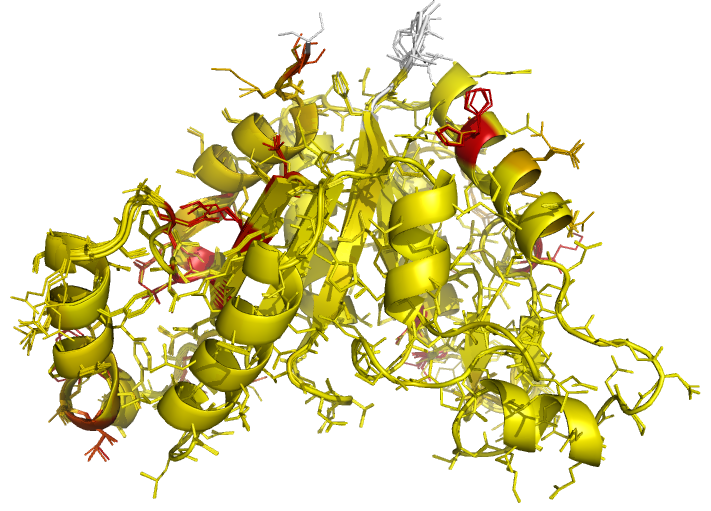
Figure 5 (stereo). Illustrations of superposed
NCS-related chains in the structure of BioD with PDB
code 3MLE (Porebski et al., 2012). Residues are coloured according to side-chain RMSD relative to the local
coordinate frame, allowing easy visual identification of residues with
side-chains in different conformations. This can be particularly useful for
cross-validation during various stages of the refinement process (e.g. by
identifying changes in side-chain conformation before/after refinement, and
identifying which side-chains are/aren't pulled towards reference structures
after application of external side-chain restraints).
External
Restraints for use in Macromolecular Crystallographic Refinement
Information from well-refined higher-resolution
structures may be used to improve reliability of low-resolution structures
during refinement, provided that the reference structure is sufficiently
similar to the target. When using such external information, there are various questions
one should ask that might affect restraint generation, for example:
- How structurally similar is the
external reference structure to the target structure being refined?
- Are the structures identical in
sequence?
- Is the reference structure
high-resolution and well refined? Should it be re-refined?
- Does it make sense to restrain
side-chain atoms?
- If only main-chain restraints are
to be used, is the backbone of the reference
structure sufficiently well refined, compared with that of the target
structure, in order for the target structure to benefit from the use of
external restraints?
- If there is not a suitable
reference structure, is it possible to generate generic restraints instead
(e.g. for alpha-helices)?
Here, we discuss how ProSMART can
be used to generate interatomic distance restraints
for subsequent use in refinement by REFMAC5. Details of the methods used, along
with examples of application, are described in an upcoming article (Nicholls et
al., 2012).
Choice of external reference structure(s)
External reference structures would usually be identical
or close homologues, although in theory any structures could be used. There are
no hard-coded limits on sequence identity, although intuitively only
sufficiently similar structures should be used as external information.
However, ProSMART is general and flexible, and
provides the ability to allow user-input beyond the realms of common sense!
It is possible to use ProSMART
to generate restraints using multiple reference structures. If multiple
homologues are provided then REFMAC automatically selects the regions that are
most consistent with the existing structure, only using the restraints that are
closest to the current interatomic distances. This is
done separately for each interatomic restraint. This
means that if many structures are used as references, only the structure(s)
most similar to the target should actually affect refinement, in theory.
Nevertheless, we do not suggest that blindly using all homologues is a good
strategy - manual consideration and common sense should always prevail!
Carefully selecting one or a selection of homologous structures that are highly
conserved in local structure would generally be a better approach, at present.
Note that the target and reference structures should be
conserved in local structure - conservation of overall fold at the
global level is neither necessary nor sufficient. To clarify, the
conformation-independent approach of ProSMART means
that sensible external restraints may be generated even if the target and
reference structures adopt different global conformations (e.g. apo and holo forms). Indeed, the
external restraints generated by ProSMART always
operate locally, and thus they should not enforce global rigidity.
If a reference structure contains multiple (e.g.
NCS-related / multimeric) chains in the PDB file then
ProSMART will generate restraints for all target
chains using information from all reference chains, by default. This may be
desirable in some cases, for reasons outlined above. However, it is also
possible to avoid this, and instead generate restraints only for the chain
considered to be the best match to the target chain, in terms of net local
structural similarity (keyword: -restrain_best).
It is important not to forget that reference structures
may contain errors - the naïve application of restraints from such
structures may cause errors to propagate into the target structure during
refinement. Consequently, it is recommended to inspect reference structures,
and possibly also consider their re-refinement before use, e.g. using PDB_REDO
(Joosten et al., 2009).
Selection of suitable structural information
It is recommended to perform a comparative structural
analysis between target and reference structures prior to refinement (i.e.
viewing the results from ProSMART ALIGN). This allows
the user to visualise and quantify local (dis)similarities between the structures and thus make
better-informed decisions regarding the local structural similarity of their
presumed homologue to the target, prior to the application of the external
restraints. It is also recommended for such comparative analyses to be
performed following refinement, allowing the user to visualise
and quantify the effect of the external restraints on local structure. In
tandem with inspection of the electron density, this would help in deciding
whether the external restraints were constructive (thus should be kept) or
destructive (thus should be removed or replaced) in each local region, and also
help in deciding the values of REFMAC5 external restraint weighting parameters
(see below).
Ideally, the target and reference structures should be
manually inspected, and the decision should be made as to whether restraints
should or should not be generated for all regions. Challenging structures may
require special attention, however, this should be deemed worth the effort when
the alternative is to produce a better diffracting crystal. If there are some
regions that are actually different between the two structures, and it is
decided that external restraints should not be generated for these regions,
then the restraints corresponding to these residues/regions can be removed
(keywords: -restrain and -restrain_rm).
Additionally, it is possible to specify that restraints should only be
generated for regions that are sufficiently conserved, in terms of local
main-chain (keyword: -cutoff) and/or side-chain (keyword: -side_cutoff)
similarity. By default, restraints are generated for all aligned portions of
structure, regardless of local structural conservation - the user must decide
whether dissimilarity thresholds are appropriate in the particular case.
Alternatively, PDB files may be separately filtered in a way deemed appropriate
for subsequent restraint generation, either manually or using a tool such as
Tim Grüne's mrprep (Grüne, 2012).
Note that restraints can be generated only for main-chain
chain atoms (keyword: -main), or for both main-chain and side-chain atoms (keyword: -side).
Important REFMAC5 parameters
At present, caution is due when optimising
certain parameters in REFMAC5, most notably the external restraints weight
(keyword: EXTERNAL WEIGHT SCALE) and Geman-McClure parameter
(which down-weights outliers, keyword: EXTERNAL WEIGHT GMWT), in
order to successfully apply external restraints during refinement. For more
information, see Nicholls et al. (2012). Information on how to provide
REFMAC5 with such keywords can be found here and here; they can also be specified using the CCP4i REFMAC5
GUI (see below).
Appropriate values for these parameters will vary for
each case, and will depend on many factors. Such factors may include properties
of the target structure's refinement in the absence of external restraints
(e.g. X-ray resolution, quality of electron density, geometry weight, number of
NCS-restrained chains) and also properties of the external structural
information, such as the quality of the external information (i.e. reference
structures), and the type of external restraints (e.g. main-chain, side-chain,
generic SS H-bond or fragment-based restraints), etc.
As an aside, it is important to make sure that sufficient
cycles of REFMAC5 are executed when using external restraints - the external
restraints will seem to have little effect if running only 5 refinement cycles.
Something like 20-30 cycles may be required, and even more if also using
jelly-body restraints (Murshudov et al., 2011).
Generic fragment and secondary-structure-based restraints
Further to generating restraints using homologous protein
structures, ProSMART allows the generation of
restraints to specific n-residue structural fragments (keyword: -lib to use
all fragments present in the local library). Such generic fragment-based
restraints can be used to help the structure better-adopt a desired
conformation, for example, using an ideal helical fragment to help stabilise the formation of a helix. The implemented
approach is generalised, allowing any structures to
be used as reference fragments in principle (e.g. the user may provide their
own fragment coordinate files). At present, ProSMART
comes with fragments representing an ideal alpha-helix (keyword: -helix) and a
representative beta-strand (keyword: -strand), which
can be used to generate in-sequence quasi-secondary-structure restraints. Note
that these restraints are not considered true secondary-structure restraints,
since they restrain all atom-pairs in the local structural environment, rather
than just those considered to be hydrogen-bonded (also, note that the residue
alignment is not assigned using a traditional method such as DSSP). This method
is considered powerful, since it does not require the low-resolution structure
to be sufficiently well modelled to assign
secondary-structure using hydrogen-bonding patterns. For example, you may want
to use helix restraints from ProSMART to force a
particular troublesome helix to maintain a reasonable helical conformation
during early/intermediate stages of model building/refinement.
Further to existing functionalities, generic restraints
representing specific atomic interactions, notably hydrogen bonds (e.g. in
alpha-helices, across beta-sheets), and also external restraints for DNA/RNA
from homologous structures, will be available imminently in an upcoming version
of ProSMART (which should be available by the time
you read this!).
How to Run ProSMART
Please contact the authors for any assistance or
information (see here).
Command line
Basic instructions on how to run ProSMART
using a command line interface can be found here.
Typing prosmart will
give a list of all keywords in that version. A list of keywords with more
detailed descriptions can be found here. Note that keywords can be specified in an external
text file (keyword: -f), which can be useful if the arguments list gets too
long.
Instructions on how to use a command line interface to
run REFMAC5 with external restraints generated by ProSMART
can be found here.
CCP4i REFMAC5 GUI
An updated version of the REFMAC5 CCP4i GUI, which
includes the ability to automatically run ProSMART
for external restraint generation, has been developed by Martyn Winn at STFC
Daresbury. The updated interface is currently available, and will be
distributed in the next release of the CCP4 suite (version 6.3). The REFMAC5
GUI allows you to specify for ProSMART to be
automatically run (using mainly default settings) prior to running REFMAC5 with
the generated external restraints. Note that this new feature will be hidden if
ProSMART is not installed.
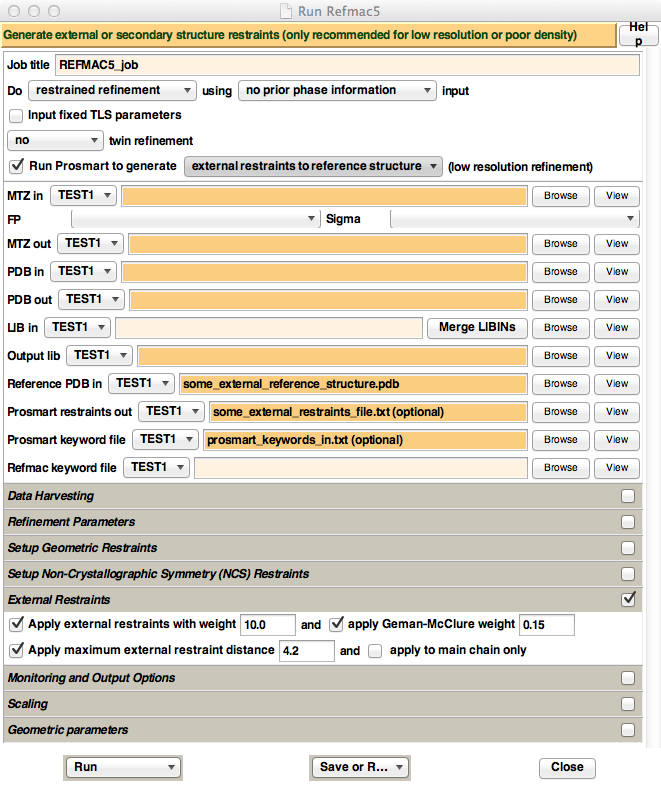
Figure 6. Appearance of the new
REFMAC5 CCP4i GUI, when ProSMART is installed. For
illustrative purposes only, text has been inputted into the "Reference PDB
in", "Prosmart restraints out", and
"Prosmart keyword file" fields - these are
the three fields that appear when enabling the "Run Prosmart
to generate" option. Also, the "External Restraints" tab has
been opened, which contains options that control the way REFMAC5 uses the
external restraints generated by ProSMART.
To automatically generate and use ProSMART
external restraints, make sure that the "Run Prosmart
to generate" button is ticked. This will cause three input file
options to appear: "Reference PDB in" (analogous to the -p2
keyword), "Prosmart restraints out",
and "Prosmart keyword file"
(analogous to the -f keyword). For simple execution, only the "Reference
PDB in" has to be specified. For more advanced functionality, create a
simple text file containing the desired ProSMART
keywords, and pass this file to the GUI in the "Prosmart
keyword file" field.
The REFMAC5 GUI has four options controlling behaviour of the external restraints during refinement,
namely the external restraints weight, Geman-McClure
parameter, maximum external restraints distance, and whether or not side-chain
atoms are to be restrained in addition to main-chain atoms. These options can
be found in the "External Restraints" tab in the GUI. Unless
there is a very good reason for doing otherwise (or just want to experiment!),
it is highly recommended to enable "Apply maximum external restraint
distance", and set it to 4.2 (which is a magic number that tends to
always be approximately optimal). Other parameters should be experimented with;
see Nicholls et al. (2012) for more information.
As an aside, it may be advisable to select "Run&View Com File" instead of "Run"
from the bottom-left drop-down box. This will display the command (with
keywords) used to run REFMAC5 before actual execution - this will confirm
whether REFMAC5 is being run as intended!
How to ensure that the external restraints are being used
by REFMAC5 during refinement
To ensure that CCP4i is running ProSMART
and REFMAC5 as intended, it is recommended to inspect the output log file to
see exactly what command was used to run ProSMART,
and confirm that the ProSMART job completed
successfully. The log file can be accessed from the main CCP4i GUI by
double-clicking the appropriate job (or alternatively selecting "View
Files from Job" then "View Log File"). For example,
at the top of this log file you should see something like this:
**************************************************************************** Information from CCP4Interface script*************************************************************************** *** Starting Prosmart to determine restraints to external structure *** Using command: prosmart -p1 "some_pdb.pdb" -o "some_location" -side -p2 "another_pdb.pdb" Writing results to directory "some_directory" ***************************************************************************This is
then followed by the main ProSMART
log. Upon successful completion, the following lines (or similar) will be
displayed after the ProSMART log:
**************************************************************************** Information from CCP4Interface script*************************************************************************** *** Prosmart finished *** Copying file of restraints some_file.txt to another_file.txt ***************************************************************************To confirm that the external restraints are being used,
locate the restraints table in the log file (just before refinement cycle 1),
which will look something like this:
--------------------------------------- Standard External All Bonds: 7711 22212 29923 Angles: 13788 0 13788 Chirals: 706 0 706 Planes: 1372 0 1372 Torsions: 3296 0 3296---------------------------------------Usage of
the external restraints is confirmed by the fact that the number of external
bonds is non-zero (and generally quite large - in this case, 22212).
As of
REFMAC5 version 5.7.0022, more output regarding external restraints will be
available in the log file, providing information about the input parameters and
options used. This information will look something like this (located above the
standard restraints table):
-------------------------------------------------------------------------------- External restraints group : 1 External restraints file :input_keywords Fail if one of the atoms involved in the restraints is missing in the pdb file Use restraints for all defined atoms Ignore restraints if abs(dmod-drest) > 50.000000 *sigma Ignore retraints if input dist > 1.00000003E+32 Weight scale sigmas : 1.0000000 Weight min sigma : 0.0000000 Weight max sigma : 100.00000 GM parameter : 0.10000000 Number of distances : 10809 Number of angles : 0 Number of torsions : 0 Number of planes : 0 Number of chirals : 0 Number of intervals : 0--------------------------------------------------------------------------------How to use the
REFMAC5 GUI with a pre-generated ProSMART
restraints file
It is
often desirable to run ProSMART separately (e.g. from
the command line, or the ProSMART GUI) instead of
running ProSMART automatically using the REFMAC5 GUI.
In this case, it is necessary to provide the REFMAC5 GUI with the external
restraints file generated by ProSMART. This
restraints file should be provided to the GUI in the "Refmac keyword file" field. Note that, in this
case, the "Run Prosmart to generate"
button should not be ticked.
For more
control over how REFMAC5 deals with the external restraints, it is advised to
create a REFMAC5 external keywords file that specifies the location of the ProSMART restraints file, and then
pass this keywords file to the GUI using the "Refmac
keyword file" field (latest versions of REFMAC5 only). To do this,
create a simple .txt file (note: must be plain text, not rich text) that
contains the commands to tell REFMAC5 to use the ProSMART
restraints file. For example, this file may simply contain the line:
@my_prosmart_restraints_file.txt
where my_prosmart_restraints_file.txt is the
name/location of the restraints file generated by ProSMART.
The @
symbol specifies for REFMAC5 to parse the my_prosmart_restraints_file.txt file and
use any external restraints found.
In
practical application, it is necessary to tell REFMAC5 how to deal with these
restraints, e.g. what weighting parameters to use. This can be achieved by
specifying EXTERNAL keywords. For example, the options enabled in the "External
Restraints" tab in the GUI shown in Figure 6 would be specified:
EXTERNAL DMAX 4.2
EXTERNAL WEIGHT SCALE 10
EXTERNAL WEIGHT GMWT 0.15
@my_prosmart_restraints_file.txt
Note that
any EXTERNAL keywords
must be specified above the @my_prosmart_restraints_file.txt line.
In
addition, the EXTERNAL USE MAIN keyword can be used to discard any side-chain restraints
that may be present in the external restraints file (this equivalent to the "apply
to main chain only" option in the REFMAC5 GUI). Of course, regardless
of whether this keyword is specified, side-chain atoms will only be restrained
if you tell ProSMART to generate side-chain
restraints (keyword: -side). The EXTERNAL USE ALL keyword may be
specified to ensure that all external restraints present in the ProSMART restraints file are used.
As of
REFMAC5 version 5.7.0022, an additional keyword EXTERNAL USE HBOND may be
specified, which only uses external restraints that may form hydrogen bonds (i.e.
restrained atom-pair are donor and acceptor); all other restraints present in
the input file will be ignored. Note that only one of EXTERNAL USE MAIN, EXTERNAL USE ALL, and EXTERNAL USE HBOND may be
specified (per external restraints file).
For clarification
of the format of REFMAC5 keywords files, here is an example keyword file.
CCP4i ProSMART GUI
A ProSMART GUI for CCP4i is available from the Murshudov
Group website (under Software→ProSMART→Download
ProSMART). Installation instructions can be found here. This GUI provides a user-friendly alternative to
running ProSMART from the command line, providing
near-comprehensive functionality for both ProSMART
ALIGN (structural comparison) and ProSMART RESTRAIN
(external restraint generation) features. Note that restraints generated using
this GUI can be passed to the REFMAC5 GUI via an external keywords file, as
described above.
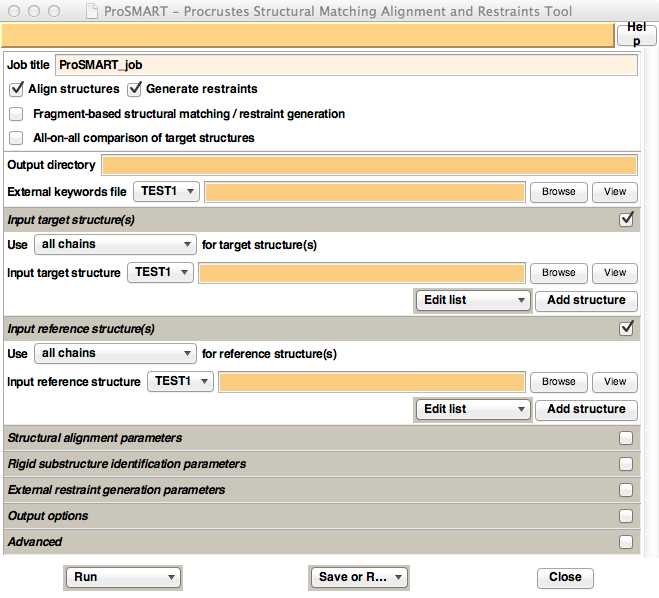
Figure 7. Appearance of the ProSMART
CCP4i GUI.
ProSMART Output
After
running ProSMART, there are two major sources of
general results information: the main ProSMART
log file, and the ProSMART HTML output page.
The main ProSMART log file indicates what
options were used (e.g. whether or not side chain restraints were generated),
which chain-pairs were considered, and whether the job finished successfully.
In the CCP4i GUIs, this log file can be accessed by double-clicking the
appropriate job (or alternatively selecting "View Files from Job"
then "View Log File"). If using a command line interface, this
information is printed to screen.
The ProSMART HTML output page allows navigation of all major
results files, provides an easy way of viewing the log files from all pairwise executions of ProSMART
ALIGN and ProSMART RESTRAIN, and provides a list of
the program options used (which can be useful for future reference). The HTML
results page may be accessed from the ProSMART CCP4i
GUI under "View Files from Job". Otherwise, this results file
may be found in the ProSMART
output directory (which by default would be here: ProSMART_Output/ProSMART_Results.html relative
to the current working directory).
The major
output files generated by ProSMART that can currently
be viewed from the HTML output page include:
- Residue-based alignment, including
various local conformation-independent structural dissimilarity scores;
- Global alignment score matrices,
which include measures that are independent of global conformation;
- Transformations required to
superpose structures, based on both global alignment and rigid
substructure identification features;
- PDB-format files containing the
structures in the coordinate frames of the (potentially various) superpositions;
- Colour scripts, used for visualisation of residue-based dissimilarity scores
and rigid substructure belongingness;
- Interatomic distance restraint files, both
for individual chain-pairs, and concatenated for all chains.
The
output superposed PDB files may be viewed using PyMOL
(Schrödinger; DeLano, 2002). The output PyMOL colour scripts may be used
to colour residues according to their various
dissimilarity scores (including those illustrated in Figures 2-5).
The
latest release of CCP4mg (McNicholas et al., 2011) includes an
experimental ProSMART analysis feature that allows
results from a ProSMART execution to be loaded for a
powerful interactive illustration of the comparative analyses. Unlike with PyMOL, CCP4mg does not use/require ProSMART
to provide superposed PDB files and colour scripts in
order to achieve the desired effect. The ProSMART
transformation files are used to superpose both global alignments and any
identified rigid substructures; these structures may be coloured
according to their residue-based dissimilarity scores, with colour
gradients altered in real-time using a slider.
Availability
and Contact Information
ProSMART will be distributed as part of the CCP4 suite (Winn et
al., 2011) in the upcoming release (version 6.3). Latest versions, along
with simple installation instructions and documentation, are always available from
the Murshudov Group website: http://www2.mrc-lmb.cam.ac.uk/groups/murshudov/
(under Software→ProSMART).
For more
information, please contact: nicholls@mrc-lmb.cam.ac.uk.
Any comments or questions are always welcome!
This
article may be freely cited and referenced, although it is preferred that
references to restraint generation using ProSMART be
made to Nicholls et al., 2012.
Acknowledgements
We would
like to thank Martyn Winn for developing the REFMAC5 GUI, Stuart McNicholas for
working on the ProSMART analysis features in CCP4mg,
Marcin Wojdyr and Charles Ballard for providing advice on software and
installation issues and working on the integration of ProSMART
into CCP4, and CCP4 for support and distribution. We would also like to thank
our colleagues for interesting discussions, and the many users who have
provided useful feedback, resulting in greatly improved functionality.
This work
was supported by the Medical Research Council (grant number: MC US A025 0104).
Part of this work was carried out whilst the authors were at the Structural
Biology Laboratory, Department of Chemistry, University of York, during which
time RAN was funded by a BBSRC Ph.D. Studentship, MF was funded by a Wild Fund
Scholarship and a BBSRC Ph.D. Studentship, and GNM was funded by the Wellcome Trust.
References
Catell, R.B. and Hurley, J.R. (1962) The Procrustes
program in producing direct rotation to test a hypothesized factor structure. Behavioural Science 7, 258-262.
DeLano, W.L. (2002) The PyMOL Molecular Graphics System.
Fischer
M., Zhang Q.Y., Hubbard, R.E. and Thomas G.H. (2010) Caught in a TRAP:
substrate-binding proteins in secondary transport. Trends in Microbiology
18(10), 471-478.
Gower, J.C. (2010) Procrustes methods. Wiley
Interdisciplinary Reviews: Computational Statistics 2(4), 503-508.
Gower, J.C. and Dijksterhuis, G.B. (2004) Procrustes problems. Oxford
University Press, USA.
Grüne, T.
(2012) mrprep - PDB preparation tool for use with ProSmart or for Molecular Replacement. http://shelx.uni-ac.gwdg.de/~tg/research/programs/mrprep/
Joosten,
R.P., Salzemann, J., Bloch, V., Stockinger,
H., Berglund, A.C., Blanchet, C., Bongcam-Rudloff,
E., Combet, C., Da Costa,
A.L., Deleage, G., Diarena,
M., Fabbretti, R., Fettahi,
G., Flegel, V., Gisel, A., Kasam, V., Kervinen, T., Korpelainen, E., Mattila, K., Pagni, M., Reichstadt, M.,
Breton, V., Tickle, I.J., Vriend, G. (2009) PDB_REDO:
automated re-refinement of X-ray structure models in the PDB. Journal of
Applied Crystallography 42(3), 376-384.
McNicholas,
S., Potterton, E., Wilson, K. S. and Noble, M. E. M. (2011) Presenting
your structures: the CCP4mg molecular-graphics software. Acta Crystallographica
D67, 386--394.
Murshudov
G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner
R.A., Nicholls R.A., Winn M.D., Long F. and Vagin A.A. (2011) REFMAC5 for the
Refinement of Macromolecular Crystal Structures. Acta
Crystallographica D67, 355-367.
Murshudov G.N., Vagin A.A. and Dodson E.J. (1997) Refinement of
Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallographica D53,
240-255.
Nicholls R.A. (2011) Conformation-Independent Comparison of Protein
Structures. University of York (thesis). http://etheses.whiterose.ac.uk/2120/.
Nicholls R.A., Long F. and Murshudov G.N. (2012) Low resolution refinement
tools in REFMAC5. Acta Crystallographica
D68, 404-417.
Porebski P.J., Klimecka M., Chruszcz M., Nicholls R.A., Murzyn
K., Cuff M.E., Xu X., Cymborowski
M., Murshudov G.N., Savchenko A., Edwards A. and
Minor W. (2012) Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals
differences between family members. FEBS Journal.
Potterton
E., Briggs P., Turkenburg M. and Dodson E. (2003) A
graphical user interface to the CCP4 program suite. Acta
Crystallographica D59, 1131-1137.
Schrödinger,
LLC. The PyMOL Molecular Graphics System, Version
1.5.0.1.
Taleb, N.N. (2010) The bed of Procrustes: philosophical and practical aphorisms. Random House.
Winn,
M.D., Ballard, C.C., Cowtan, K.D., Dodson, E.J., Emsley, P., Evans, P.R.,
Keegan, R.M., Krissinel, E.B., Leslie, A.G.W., McCoy, A., McNicholas, S.J.,
Murshudov, G.N., Pannu, N.S., Potterton, E.A., Powell, H.R., Read, R.J., Vagin,
A. and Wilson, K.S. (2011) Overview of the CCP4 suite and current developments.
Acta Crystallographica D67,
235-242.