Maximum-Likelihood Refinement of Atomic Models using Least-Squares Criterion
by
P. Afonine$,*, V.Y. Lunin#,* & A. Urzhumtsev*
$ Centre Charles Hermite, LORIA, Villers-lès-Nancy, 54602 France
# IMPB, Russian Academy of Sciences, Pushchino, 142290, Moscow Region,
Russia
* LCM3B, UPRESA 7036 CNRS, Université Henri Poincaré, Nancy 1, B.P. 239,
Faculté des Sciences, Vandoeuvre-lès-Nancy, 54506 France
e-mail:
sacha@lcm3b.uhp-nancy.fr
1. Notation
Fobs(s) -
observed structure factors magnitudes
F*(s) - modified structure
factors magnitudes
Fmod(s) -
magnitudes of structure factors calculated from the model
w(s) - weights for
least-squares terms
LS - least-squares criterion calculated with Fobs
LS* - least-squares criterion calculated with F*
ML - maximum likelihood criterion (logarithm of likelihood gain)
a, b -
parameters of the join probability distribution of structure factors considered
as a
function of random
atomic models
e - reflection multiplicity
I0(x), I1(x), I2(x) - modified Bessel functions of 0, 1 and 2 order of argument x
cosh(x), tanh(x) - hyperbolic cosine and tangent of argument x
2. Least-squares and maximum likelihood criteria
The basic goal of a crystallographic refinement is to find an atomic model such that it minimises the functional
where
the crystallographic criterion RX
describes the quality of fit of structure factor magnitudes, Fmod, calculated from the
model, to the experimental data, Fobs,
and the RO embodies other
terms such as stereochemical criteria, a phasing criterion etc. In order to
analyse the dependence of the refinement results on the choice of the
crystallographic criterion RX,
in the current work the term RO
was excluded from all calculations.
In practice, the basic statistical
hypotheses for this criterion break when the atomic model is incomplete and the
errors in experimental data Fobs
are not independent, and the LS
criterion becomes inadequate to the situation.
Recently, the maximum likelihood criterion started to be used (Bricogne
& Irwin, 1996; Pannu & Read, 1996; Murshudov et al., 1997) as RX .
The maximisation of likelihood is equivalent to minimisation of negative
logarithm likelihood gain, which may be calculated as (Lunin & Skovoroda,
1995)
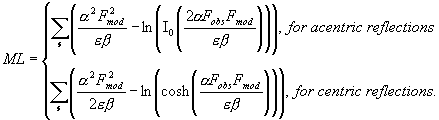 |
One of its major advantages is that it takes
into account the contribution of atoms missed in an available atomic model
(Lunin & Urzhumtsev, 1999).
However, an implementation of the ML criterion in existing programs needs
their essential modification. An alternative solution would be to approximate
this criterion near its minimum by a functional quadratic with respect to
structure factor magnitudes, calculated from the atomic model (Lunin &
Urzhumtsev, 1999). In this case, such approximation can be written again in the
form of the usual LS criterion:
The values F*
can be considered as modified magnitudes Fobs
and can be obtained as the solution of the following equation with respect to F*:
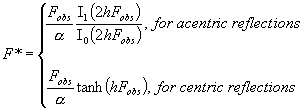 |
with a and b estimated as in (Lunin & Skovoroda, 1995)
and
The weights w*
are calculated as
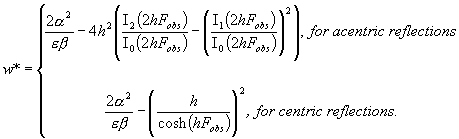 |
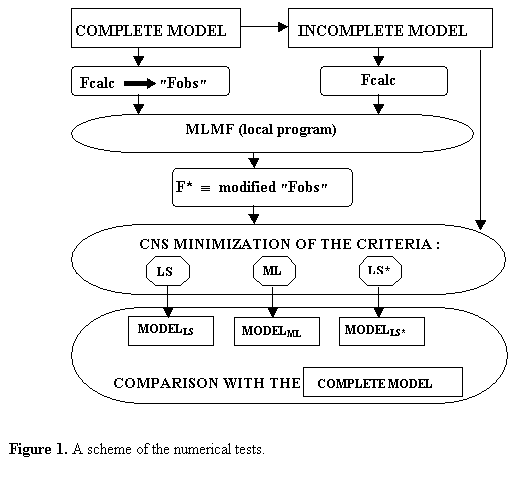
The tests below show the comparison of the
refinement with different criteria: LS,
ML and LS*. Complete tests results and their analysis will be published
elsewhere.
3. Numerical tests on comparative analysis of different criteria
The refinement tests were carried
out with CNS complex (Brünger et al.,
1998) using the structure of Fab fragment of monoclonal antibody (Fokine et al., 2000). This model includes 439
amino acid residues and 213 water molecules. The molecule crystallises in the
space group P212121 with the
unit cell parameters a = 72.24 Å, b = 72.01 Å, c = 86.99 Å, one molecule per
asymmetric unit.
For test purposes the experimental
data Fobs at 2.2 Å
resolution were simulated by the corresponding values calculated from the
complete exact model (Fig. 1). In all tests described below the starting atomic
parameters were exact. Due to the absence of some atoms, removed randomly, the
minimisation of the crystallographic criterion shifted the atomic parameters
from their exact values showing that the minimum of all these criteria does not
correspond to the correct model any longer. Smaller resulting errors indicate
better quality of the criterion.
3.1. Test1: Random deletion of atoms in the crystal
In this test the atoms were removed
randomly, the percentage of removed atoms varied from 0 to 20%. For each
incomplete models the minimisation procedure was carried out using three
different crystallographic criteria: LS,
ML and LS* (we remind that all stereochemical criteria were excluded from
this refinement).
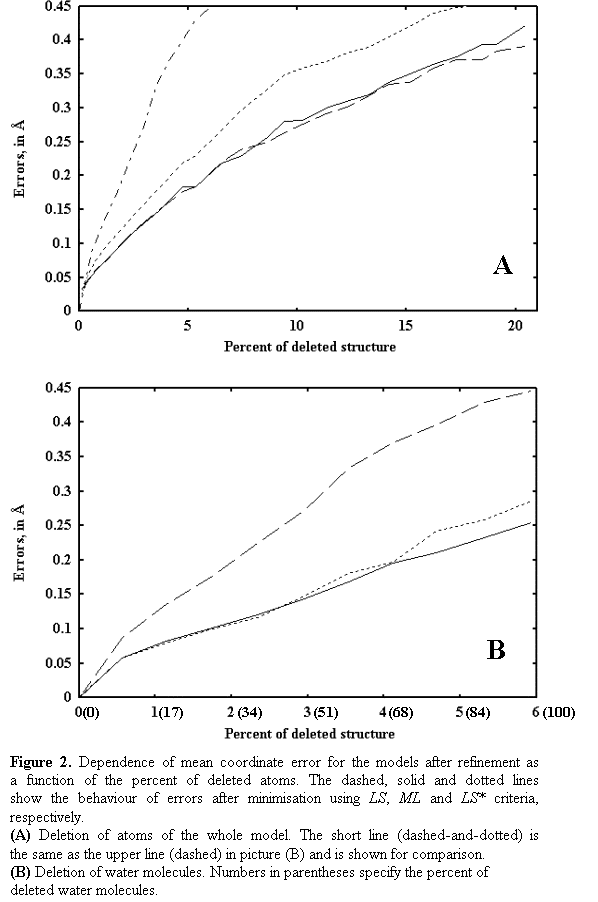
Figure 2a shows, for every
criterion, the mean error in atomic positions for the models after refinement
as a function of the size of a deletion. The errors grow with the percent of a
deleted structure. The errors obtained with the LS* minimisation are systematically less then those for the LS minimisation and are almost equal to
the errors obtained with the ML minimisation.
It can be noted that the weights w*
are crucial in order to obtain such results.
3.2. Test2: Random deletion of water molecules only
This test is similar to the previous
one with the difference that only water molecules were allowed to be deleted
from the model. The behaviour of errors (see Fig. 2b) is similar to that in the
previous case. However, these errors are significantly larger and they grow
faster with the percentage of the deleted structure (compare Fig. 2a and Fig.
2b). The reason for this may be the following: the water molecules are situated
at the surface of the protein and not in its volume, and when the same amount
of atoms is randomly excluded in both tests, in the case of water molecules
they are distributed less uniformly in the space making stronger influence on
the structure factors.
4. Conclusions
The tests discussed above show that
the incompleteness of the model can seriously affect to the refinement. The
more atoms are deleted, the larger are the errors in the model which fits best
to the experimental data. Removal of water molecules has a stronger effect than
a removal of a similar quantity of atoms randomly in the whole unit cell.
The tests show that the ML criterion is less sensible to the
absence of a part of a model than the traditional LS criterion. In the case when an insertion of the ML criterion into an existing program is
complicated, it can be replaced by its quadratic approximation. This
approximation corresponds to the LS
criterion calculated with Fobs
substituted by F* values and weighted
by w* (expression for both is given
in the text). In all tests the least-squares minimisation against modified
structure factors F* gave the models
of a significantly higher quality than those obtained by the minimisation
against simulated Fobs and
practically coinciding with the models obtained by maximum likelihood
minimisation.
This shows that any crystallographic
refinement program based on the minimisation of the least-squares criterion can
give the results of the same superior quality as using maximum likelihood
criterion without modifying the program itself when proper magnitudes and
weights are used.
In this article we presented the
results of first tests with an incomplete model without errors. An influence of
other sources of imperfection of the model and data on refinement with various
criteria will be discussed elsewhere.
The authors thank T. Skovoroda for
her help with programming and C. Lecomte for his support of the project.
References
Bricogne, G. & Irwin, J. (1996).
Proceedings of the CCP4 Study Weekend,
85-92.
Brünger,
A.T., Adams, P.D., Clore, G.M., DeLabo, W.L., Gros, P., Grosse-Kunstleve, R.W.,
Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N.S., Read, R.J., Rice, L.M.,
Simonson, T. & Warren, G.L. (1998) Acta
Cryst., D54, 905-921.
Fokine, A.V., Afonine, P.V., Mikhailova, I.Yu., Tsygannik, I.N.,
Mareeva, T.Yu., Nesmeyanov, V.A., Pangborn, W., Li, N., Duax, W., Siszak, E.,
Pletnev, V.Z. (2000). Rus. J Bioorgan
Chem, 26, 512-519.
Lunin,
V.Y. & Urzhumtsev, A.G. (1999). CCP4
Newsletter on Protein Crystallography, 37,
14-28.
Lunin,
V.Y. & Skovoroda, T.P. (1995). Acta
Cryst., A51, 880-887.
Murshudov,
G.N., Vagin, A.A. & Dodson, E.J. (1997). Acta Cryst., D53,
240-255.
Pannu,
N.S. & Read, R.J. (1996). Proceedings
of the CCP4 Study Weekend, 75-84.
 |
Newsletter contents...