Maurice E. Müller Institute for Microscopic Structural Biology
Biozentrum, University of Basel
Klingelbergstrasse 70
4056 Basel, Switzerland
Almost every software package for crystallography and image processing defines its own image file format, creating unending headaches for the user. Standardization is a useful approach to lessen the administrative problems associated with format conversion. However, this only remains a practical solution as long as the field remains isolated. In contrast, modern science drives towards large-scale integration of data obtained from diverse sources, requiring ever more conversion between different file formats.
The traditional solution to the conversion problem is a giant program, converting an input file format into a common one, before writing the desired format. While this is a viable approach widely used, it means having only this one program able to deal with all the different formats. In addition, this leads to the proliferation of copies of a data set that needs to be kept in different formats. Furthermore, the programmer writing data processing code must then decide which file format he likes, and often he invents a new format as well.
An effort to rationalize the integration of scientific data from different sources, especially microscopic image data, developed into a database prototype project called BioImage. One of the issues that had to be resolved, was which file format or formats would be adopted. It was clear that some sort of conversion utility would be needed for a variety of file formats, including the crystallographic formats, CCP4 and MRC, as well as other formats developed for single particle analysis by electron microscopy (such as SPIDER) and confocal light microscopy (such as BioRad). To write yet another conversion program in the conventional style seemed to be a poor solution.
Because the BioImage project also involves the development of 3D visualization in collaboration with SGI (Silicon Graphics Inc.) [1], it was logical to look at the way file format handling is done on SGI computers. It turned out that SGI's Image Format Library (IFL) provides a convenient way of specifying file formats once for multiple uses, even in existing programs not specifically written for some particular formats. The library allows specification of a large number of image properties or attributes, providing a rich set of options to deal with existing and new file formats. This is therefore a good basis for more specialized formats such as those for molecular structures and crystallography.
The ease with which new formats can be specified in IFL was and is exploited to provide extensions covering the relevant crystallographic and microscopic image formats. The focus of these extensions was originally macromolecular 2D and 3D data sets, and therefore the set of extensions is called the "Macromolecular IFL" or MMIFL. In particular, an effort was made to provide a mechanism to preserve crystallographic and other information contained in the headers of the new file formats. Here I report on the philosophy of design and progress of MMIFL.
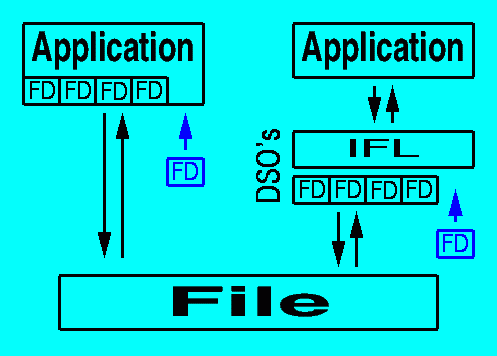
SGI's IFL (part of the ImageVision Library) provides an elegant way to deal with a proliferation of file formats without the need to recompile the source code of applications (Figure 1). The base classes of IFL deal with extracting parameters common to all image formats, thus defining a single foundation for handling all image formats. The particular features of image file formats are specified in subclasses of the generic file classes, each compiled in a DSO (Dynamically Shared Object) and placed in the appropriate library directory (/usr/lib for old 32-bit objects and /usr/lib32 for new 32-bit objects). Each file format must then be "registered" by placing a reference to it in a special file: /usr/lib/ifl/ifl_database (Figure 2). The formats already registered in this file can be listed using the program "imgformats".

Figure 1: Traditional applications (left) directly include file format definitions (FD's) within the code, thus limiting support for file formats to those the programmer included. Applications using the Image Format Library (right) have access to any format definition defined in the library (implemented as Dynamically Shared Objects on SGI computers). Addition of new format definitions (FD in blue) requires recompilation of the conventional application (left), but only the compilation and addition of a single DSO within IFL (right).
Figure 2: The registration of the CCP4 format in the file, /usr/lib/ifl/ifl_database, gives the description returned by the program "imgformats", the name of the DSO, and the suffixes identifying the file format.
The common design of crystallographic and other scientific image file formats consists of a relatively simple header of constant or varying size, followed by the 2D or 3D data. Some of the information in the header coincides with attributes defined within IFL, in particular the size, the data type (called the mode in crystallography jargon), and statistical information such as the minimum and maximum. The rest of the information need to be accessed via tag-based extensions defined in the file format.
It was decided to create one source code header file, iflMacroMol.h, for the whole group of macromolecule structure file formats. The file formats implemented in MMIFL version 0.5b are shown in Table 1. This source code header file specifies the tags required to obtain information from any of the header formats included. Each image header format is referenced as a structure which can be accessed from a C program using the tag "HEADER". Information such as the space group and unit cell parameters are associated with the tags "SPACEGROUP" and "CELL".
The extensions in MMIFL is written to conform as closely with the format specifications as possible to allow the use of converted data sets in the original software packages. Where information for particular fields in the headers is lacking, suitable defaults tailored for each file format are introduced. Conversions from little-endian and VAX-type floating point numbers are intended to be done automatically, although the detection of byte order is not trivial for all file formats. As MMIFL progresses, these issues will be addressed to provide a clean and simple interface for file format handling.
Table 1: Scientific image file formats of MMIFL
| Format | Field(s) | Description |
| CCP4 | XRD, EC | Widely used format in X-ray crystallography |
| MRC | EC, EMIP | Widely used format in electron crystallography |
| GRD | EMIP | Basel in-house format for 3D reconstructions |
| BRIX | XRD, EC, M | Map format for the program "O" |
| MFF | XRD, EC, M | Map format for the program "What If" |
| EM | EMIP | Single particle analysis package from the MPI, Martinsried |
| SPIDER | EMIP | Single particle analysis |
| IMAGIC | EMIP | Single particle analysis |
| BIORAD | CLM | Format for confocal data acquisition from BioRad |
| DI | AFM | Format for AFM dat acquisition software from Digital Instruments |
XRD: X-ray diffraction
EC: Electron crystallography
EMIP: Image processing for electron microscopy
CLM: Confocal light microscopy
AFM: Atomic force microscopy
M: Model building
IFL and MMIFL offers a set of file format definitions available to the programmer, in particular in MMIFL mechanisms to use crystallographic and other scientific information in the file headers. To illustrate its utility and provide a useful application at the same time, a simple conversion program was written called "mmcopy". The intention with this program was to be able to convert between different formats seemlessly without losing important header information. Crystallographic parameters are read if present, and written if the target file format includes them. If no such parameters are derived from the input file, defaults are generated to be written into a target file. Different file formats may support different data types, and mmcopy attempts to convert the input format into an appropriate data type for the output format.
Because of the facility with which different formats can be accessed using MMIFL through mmcopy, several manipulation options were added (Figure 3). These include explicit format changes to byte or floating point, selection from multiple images in the input file, resizing, reslicing, and changing header information. Many of these options arose from a need to prepare files for input to other programs, such as the BRIX format for O and the MRC format for image processing with the MRC package.

Figure 3: Typing "mmcopy" alone gives brief instructions for usage.
Direct access to the formats defined by MMIFL facilitated my work in electron crystallography and visualization for the BioImage project. Because a standard program on SGI computers such as imgview uses IFL, it is now a simple matter to directly look at a map (Figure 4 left). In addition, a VRML isosurfacing node developed for the BioImage project also uses IFL, allowing rendering of a map in 3D with the Cosmoplayer plug-in for Netscape (Figure 4 right). The BioImage project requires direct and automatic access to image files, and the program mmcopy has been integrated with the database prototype for converting and downloading image data. Support for interactive isosurfacing in VRML is also used to provide 3D visualization within the context of the database.
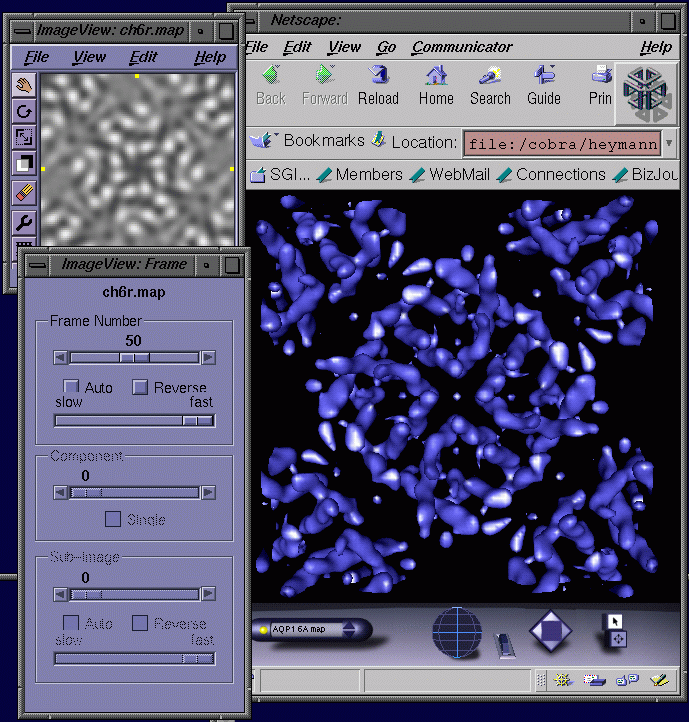
Figure 4: Specification of the CCP4 map format as part of MMIFL allows the display of maps with the program "imgview" (left) and an isosurface rendering in a VRML world (right). In imgview every frame (i.e., slice in the z-direction) can be viewed in a 3D data set. The isosurfacing in the VRML world uses a new experimental node developed in the BioImage project and is available from SGI's developers program. The map shown is that of the red blood cell water channel, aquaporin 1, obtained by electron crystallography at 6 Å resolution [2].
It is clear IFL offer a mechanism to deal with image file formats in an almost transparent way. However, its limited availability for SGI and Windows computers is a serious hurdle to widespread acceptance. Several avenues will be explored to overcome these restrictions. In the meantime, MMIFL will be further extended on SGI computers.
The MMIFL library was developed within the BioImage Project to facilitate integration of scientific and non-scientific visualization and manipulationtools into a biological image database. The extensions are limited to SGI computers and were tested on the O2 and Octane and require IRIX 6.2 and IFL 1.1.1 or later versions. Programs and utilities using IFL (such as imgview and imgcopy) can be found in the ImageVision package.
The current package is available as a gzipped and tarred file at:
http://www.mih.unibas.ch/Bioimage/mmifl_0.5.tar.gz
Documentation on MMIFL is available at:
http://www.mih.unibas.ch/Bioimage/iflMacroMol.html
Another link with a mailing list can be found at SGI's developer program site:
http://www-devprg.sgi.de/devtools/tools/MMIFL/
Acknowledgments
I wish to thank J.J.Pittet from SGI for initial support and discussions on IFL and A. Engel for his support and discussions. Work on MMIFL and the BioImage project was supported by the European Union through grant PL 960472.
References