Contents
Secondary structureSolvent Accessible Surface Area
Interfaces (Buried Area)
Hydrogen Bonds
Close contacts
 |
CCP4 Molecular Graphics Documentation | |
| Structure analysis |
| Documentation Contents | On-line Documentation | Tutorials | CCP4mg Home |
The option to colour by secondary structure is on the Colour scheme menu for any model containing protein. A Colour legend option is on the display object icon menu. Also on the display object icon menu, if the object is coloured by secondary structure, is an option to List secondary structure; the listing can then be printed to a file.
The secondary structure is assigned by the method of Kabsch and Sanders(1983). The method first finds all hydrogen bonds between main chain amide and main chain carboxy groups. Then if hbond(i,j) is a hydrogen bond between the carboxy oxygen of the ith residue and the amide group of the jth residue the secondary structure types are then defined as follows:
| 3-turn | If hbond(i,i+3) exists then residue i+1,i+2,i+3 are a 3-turn |
| 4-turn | If hbond(i,i+4) exists then residue i+1,i+2,i+3,i+4 are a 4-turn |
| 5-turn | If hbond(i,i+5) exists then residue i+1,i+2,i+3,i+4,i+5 are a 5-turn |
| alpha helix | Two or more consecutive 4-turns |
| beta strand | Two consecutive residues in one strand have hydrogen bond bridges to two consecutive residues in a neighbouring strand. |
| beta bulge | In two adjascent beta strands, two residues (called 1 and 2) are opposite one residue (called chi) in the other strand |
The beta bulge definition is taken from Richardson et al.(1978).
It is possible to override the automatic secondary structure definition: click on the model icon (the dot next to the name) and select Structure definition and Edit secondary structure to see the window:
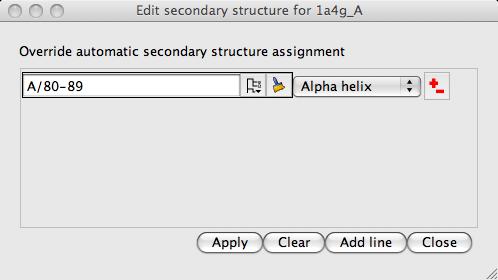
Each line in this window defines a selection of residues and the secondary structure to be assigned to that range of residues. Beware that when you run the program in future these changes will be applied automatically.
It is also possible to load the secondary structure assignment output by the DSSP program (http://swift.cmbi.ru.nl/gv/dssp/) using the Load DSSP secondary str option on the Structure definition menu.
The option to colour by solvent accessible surface (SAS) area is on the Colour scheme menu (on both Atom properties and Residue properties sub-menus). A Colour legend option is on the display object icon menu. Also on the display object icon menu, if the object is coloured by SAS, is an option to List solvent access which will list the SAS for each atom and residue, and totals for all atoms and sets of atoms specified in the Preferences. The listing can written to a file.
The SAS area is the surface area of an atom or residue which is on the surface of the molecule and therefore accessible to solvent. Conceptually the surface of the molecule is defined by rolling a sphere (notionally equivalent to a solvent molecule) over a structure made up atomic spheres with radii of van der Waals radius. The SAS is traced out by the centre of the rolling 'solvent' sphere and so this surface is further out than a surface defined by just van der Waals spheres about atoms. A macromolecular surface has many narrow indentations which are inaccessible to the rolling 'solvent' sphere and so the SAS surface tends to smooth over these narrow indentations.
CCP4mg uses the method of Lee and Richards (similar to the implementation in the CCP4 program areaimol). The SAS is calculated for the whole model with solvent molecules and hydrogen atoms excluded. Only the first of any alternate located atoms are included. The excluded atoms will be coloured in an 'out-of-range' colour.
Interface to.. indicated by colouring which atoms or residues are in contact with another set of atoms. This is equivalent to the buried area which is calculated for a set of atoms in the context of a second set of atoms. The buried area for an atom in the first set is the difference between its SAS in the presence of only the first set of atoms and in the presence of both sets of atoms.
The parameters used to calculate solvent accessibility and buried area can be changed in the Preferences window (Model analysis folder):
The water radius is conventionally set to 1.4Å which is most appropriate for comparison of results
The point density parameter determines the resolution of the calculation,the default is 1Å, but decreasing this value should improve the result but increase the calculation time.
Exclude solvent By default solvent is automatically excluded from the calculation and will be coloured in the 'below normal range' colour.
List total SAS for selected atoms There are two options to enter a selection command that will be used to select a set of atoms (by default one is 'polar_atoms'). The total SAS for all atoms in these selected set will be written out by the List solvent access command.
The default colour scheme can be changed in the Preferences window (look under Model colours).
Hydrogen bonds is one of the options on the Add display object.. sub-menu of the model icon menu. Display Table and usually hydrogen bonds are displayed for all the currently displayed atoms in the model. The first and third column in the Display Table for hydrogen bonds have two menus for selecting two sets of atoms - only the hydrogen bonds between these sets of atoms will be displayed. Note that it is possible to find hydrogen bonds to a different model: the menu in the third column has the option As display object.. which lists the display objects of the this model and all other loaded models. The hydrogen bond display object icon menu has options to change the style and labelling of the displayed hydrogen bond and options to list or print the hydrogen bonds. The hydrogen bonds can also be written out as a vectors file which can be imported and manipulated as vectors which provides more flexibility in editing and display style which may be useful when creating presentation graphics.
The parameters for calculating hydrogen bonds can be changed in the Preferences window (look under Model analysis).
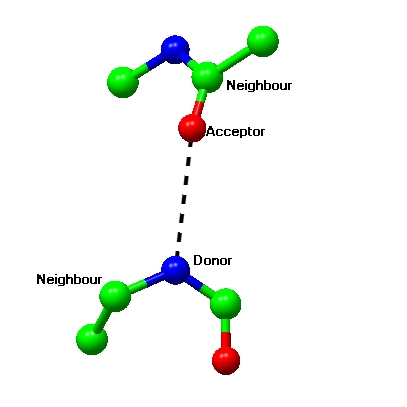
There are two sets of criteria for with and without hydrogen atoms defined. The parameters are maximum and minimum donor atom or hydrogen atom to acceptor atom distance, and minimum angles between three atoms. The default criteria are generous and will show hydrogen bonds of poor geometry.
The interface to close contacts is essentially the same as for hydrogen bonds - see above.
The search for close contacts will work in either 'simple' mode, finding all inter-atomic distances between given minimum and maximum values, or in 'van der Waals' mode, finding inter-atomic distances which are between a given minimum and maximum fraction of the sum of the van der Waals radii of the two atoms. The choice of mode and the maximum and minimum values can be changed in the Preferences window (look under Model analysis). There are also options to label close contacts with the fraction of van der Waals contact distance (rather than absolute distance) and to exclude close hydrogen-bonded atoms.
Kabsch and Sanders(1983), Dictionary of Protein Secondary Structure: Pattern Recognition of Hydrogen Bonded and Geometrical Features, Biopolymers, 22, 2577.
J.S. Richardson, E.D. Getzoff and D.C. Richardson (1978).
The Beta Bulge: A Common Small Unit of Nonrepetitive Protein Structure.
PNAS, 75, 2574-2578.>
B.Lee and F.M.Richards, (1971). J.Mol.Biol., 55, 379 - 400
E.N. Baker and R.E. Hubbard. Hydrogen bonding in globular proteins. Prog.
Biophys Mol Biol, 44:97-179, 1984.