K Cowtan
The 'nautilus' software is a new tool for automated building of RNA/DNA from electron density. It uses similar ideas to the 'buccaneer' software (Cowtan, 2008) for protein model building, but with a different and highly efficient target function for identifying nucleotide features.
The software will locate likely nucleotide features including sugars and phosphates, grow these into chains, merge overlapping chains, match the built chains to the sequence, and build the bases. The resulting structure is refined using 'refmac', and the calculation is iterated to obtain a more complete structure.
The software may be used to build nucleotide structure in experimentally phased maps, molecular replacement maps, or to add the nucleotide components to protein complexes.
The calculation consists of the following steps:
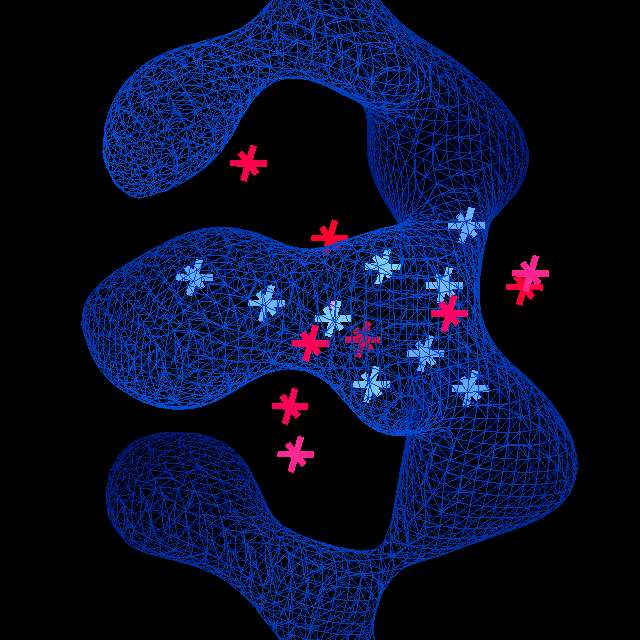
The calculation is notable for its speed, typically taking no more than a few tens of seconds. When combined with refinement in refmac (itself a fast package) (Murshudov et al, 1999), more than 90% of the time is spent in the refinement step. This speed is achieved through use a highly optimised 'fingerprint' for detecting structural features from the electron density values at a few highly informative points, which must have extreme density values if the feature is present in a given orientation. A fast rotation and translation search can be carried out using a method similar to the that employed by the ESSENS software (Kleywegt & Jones, 1997). The fingerprint for a sugar group is shown in figure 1.

Similar fingerprints are used to identify phosphates and to distinguish between different base types.
Version 0.3 of Nautilus is included in CCP4 version 6.3.0. The pipeline may be run through the CCP4i graphical user interface, or from the command line as either a build/refine pipeline or for a fast build only. In future an interactive version of some of the functionality will be available in the 'Coot' model building software (Emsley et al, 2010).
This article may be cited freely.