A self-rotation puzzle
Zhenbo Cao1,2,*, Neil W. Isaacs1
1WestCHEM, Department of Chemistry, University of Glasgow, Glasgow G12 8QQ, UK
2Department of Biochemistry and Molecular Biology, University of Glasgow, Glasgow G12 8QQ, UK
*corresponding author, E-mail: Zhenbo@chem.gla.ac.uk
Introduction
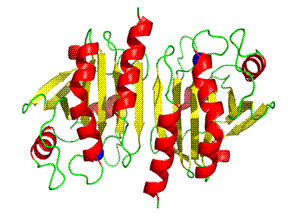
The peroxiredoxins (Prxs) are a ubiquitous family of antioxidant enzymes that regulate intracellular levels of H2O2 where they are implicated in both tissue protection against oxidative stress and H2O2-mediated signalling pathways (Wood et al. 2003). In recent years, their key role in antioxidant defence has been emphasised by their high abundance in both bacterial and mammalian cells. Peroxiredoxin III (Watabe et al. 1994) is a typical member of the 2-Cys PrxIII subclass with catalytic cysteines at its N(Cys47) and C(Cys168) termini and with a dimmer as the functional unit. Electron Microscopy (EM) studies (Gourlay et al. 2003; Wood et al. 2003) have shown that PrxIII exists as an oligomeric ring. We have determined the crystal structure of bovine mitochondrial PrxIII C168S mutant at 3.3┼ resolution (Cao et al. 2005).
What is the puzzle?
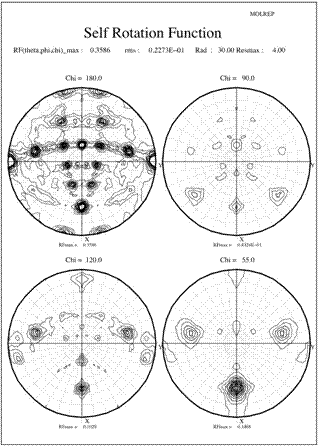
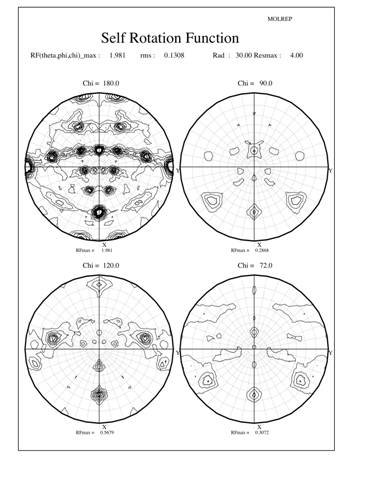
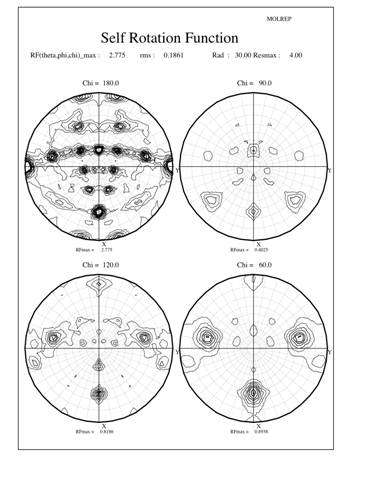
Crystals production, data collection and processing were described previously (Cao et al. 2005). The crystals belong to the monoclinic space group C2 with the Matthews coefficient (Matthews 1968) suggesting 10 (Vm=2.8 ┼3/Da) or 12 (Vm=2.3 ┼3/Da) monomeric subunits in the crystal asymmetric unit. The usual statistical indicators (CCP4 1994) gave no indication of crystal twinning. Since most (6 out of 8) known typical 2-Cys Prxs structures are decamers, a self-rotation function was calculated using the program Molrep to locate the expected NCS two-fold and five-fold axes. The results are shown in Figure 1 as stereographicprojections of polar angles. The absence of substantial peaks on the chi=72-degree section indicate there is no 5-fold symmetry in the crystal structure (Fig 1a). However, 3 peaks on the chi=60-degree section indicate three 6-fold symmetry axes, each of which is perpendicular to 6 two-fold axes shown as lines of 6 peaks in the chi=180-degree section (Fig 1b). In each line the peaks occur at an angle of about 30 degrees to each other and are perpendicular to a 6-fold peak, which also coincident with a 2-fold peak. Taken together, these facts suggest a dodecameric rather than a decameric structure. However, as the Matthews coefficient suggested only one dodecameric ring in the asymmetric unit, the indication of three different 6-fold axes of rotation really puzzled us.
Initial attempts to solve the structure by molecular replacement by MolREP (CCP4 1994), AMoRe (Navaza 1994) and PHASER (Storoni et al. 2004) with the default parameters using thioredoxin peroxidase B (TPxB) (PDB Code 1qmv) monomer, dimer and decamer as the starting model were all tried without any successful solutions. A solution was finally obtained by the molecular replacement program PHASER (Storoni et al. 2004) using the dimer of TPxB as a search model (Cao et al. 2005).
What is the answer to the puzzle?
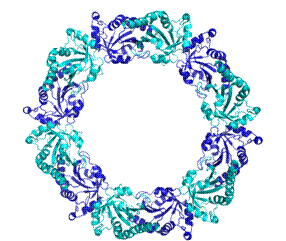
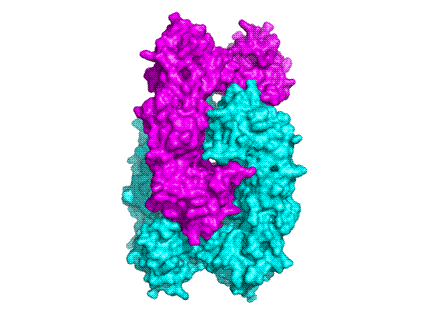
The structure shows that stable PrxIII dimers are formed across a non-crystallographic 2-fold axis that extends the central b-sheet. The 6-fold NCS-related dimers are assembled into a dodecameric ring structure with outer and inner diameters of 150 and 70 ┼ respectively (cf 130 and 60 ┼ for the decameric Prxs) (Fig 2a and 2b).
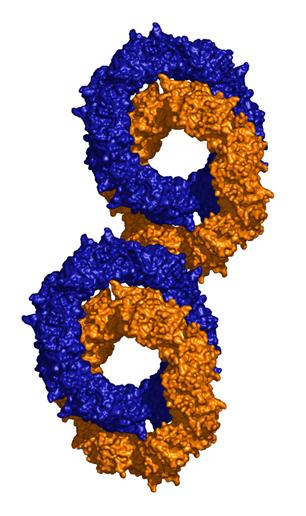
The surprising feature of the crystal structure of PrxIII C168S is its presence as a 2-ring catenane comprising two interlocking dodecameric toroids (Fig. 2c), which are arranged such that half of one ring is related to half of the other ring by the crystallographic 2-fold axis. The planes of the rings are not at right angles, but are inclined at an angle of 55░, which allows a larger contact surface between the rings. This structure provides the solution to the self-rotation puzzle. The two 6-fold peaks (chi=60) at f= 66, j=104 and f= 66, j=-104, with coincident 3-fold (chi=120) and 2-fold (chi=180) axes, are the two 6-fold axes perpendicular to the planes of the rings. The two lines of six 2-fold peaks (f= 90, j=+-165; f= 64, j=+-150; f= 38, j=+-130; f= 25, j=+-75; f= 38, j=+-23; f= 64, j=0) represent the 2-fold axes in the plane of the ring. Because the ring is composed of six homodimers, there are 12 2-fold axes in the plane, giving a 30 degree angle between adjacent axes. What is the large 6-fold, peak at f= 64, j=0?This is actually the tail of a peak arising from an improper rotation of 55 degrees (Fig 3a), which is the angle between the planes of the two rings (Fig 3b). Finally, the 2-fold peak at f= 26, j=180 is perpendicular to this and relates the two rings as shown in Fig 3b where the axis is in the plane of the page and runs vertically through the centre of the model. The two peaks at f= 90, j=90 and f= 90, j=-90 are the crystallographic 2-fold symmetry axes of the spacegroup C2.
Other catenanes
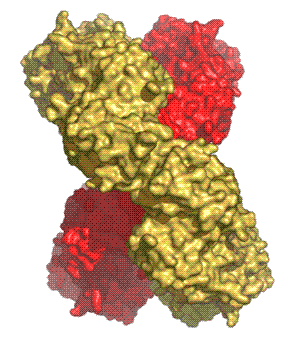
There are three previous examples of protein catenanes cited in the literature and two of them are rather specialised cases. One is a totally artificially-produced peptide catenane based on a small segment of a dimeric mutant of the p53tet protein generated in vitro using chemical techniques (Yan and Dawson 2001). Another one is a viral capsid assembly of 420 subunits where the subunits are topologically linked by covalent (isopeptide) bonds creating a form of protein Ĺchain mailĺ which is highly resistant to degradation (Wikoff et al. 2000). The third example is the crystal structure of RecR from Deinococcus radiodurans, which is involved in homologous recombinational DNA repair in procaryotes (Lee et al. 2004) (PDB ID: 1VDD). Four RecR monomers form a ring-shaped tetramer of 222 symmetry with a central hole of 30-35 ┼ diameter. In the crystal, two tetramers are concatenated (Fig 4).
How is the catenane formed?
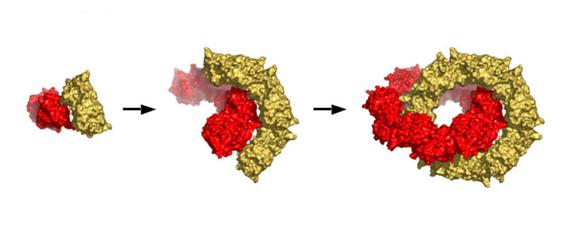
We have no data indicating how the 2-ring catenane structure is formed but a model described previously (Cao et al. 2005) is shown in Fig 5. Briefly, dimeric units can interact in two different modes that are not mutually exclusive. One mode produces the dimer-dimer contacts, primarily hydrophobic, associated with ring generation in this and other Prx structures. The other mode gives polar contacts that could potentially initiate catenane formation at any stage during single toroid assembly by allowing two rings to form simultaneously around each other.
At present it is unclear whether the catenane structure has any physiological relevance, but it provides interesting new insights into protein topology and mechanisms of subunit assembly.
References:
Cao, Z., A. W. Roszak, et al. (2005). "Bovine mitochondrial peroxiredoxin III forms a two-ring catenane." Structure (Camb) 13(11): 1661-4.
CCP4 (1994). "The CCP4 Suite - Programs for Protein Crystallography." Acta Crystallographica Section D-Biological Crystallography 50: 760-763.
Gourlay, L. J., D. Bhella, et al. (2003). "Structure-function analysis of recombinant substrate protein 22 kDa (SP-22). A mitochondrial 2-CYS peroxiredoxin organized as a decameric toroid." J Biol Chem 278(35): 32631-7.
Lee, B. I., K. H. Kim, et al. (2004). "Ring-shaped architecture of RecR: implications for its role in homologous recombinational DNA repair." Embo J 23(10): 2029-38.
Matthews, B. W. (1968). "Solvent content of protein crystals." J Mol Biol 33(2): 491-7.
Navaza, J. (1994). "Amore - an Automated Package for Molecular Replacement." Acta Crystallographica Section A 50: 157-163.
Storoni, L. C., A. J. McCoy, et al. (2004). "Likelihood-enhanced fast rotation functions." Acta Crystallogr D Biol Crystallogr 60(Pt 3): 432-8.
Watabe, S., H. Kohno, et al. (1994). "Purification and characterization of a substrate protein for mitochondrial ATP-dependent protease in bovine adrenal cortex." J Biochem (Tokyo) 115(4): 648-54.
Wikoff, W. R., L. Liljas, et al. (2000). "Topologically linked protein rings in the bacteriophage HK97 capsid." Science 289(5487): 2129-33.
Wood, Z. A., E. Schroder, et al. (2003). "Structure, mechanism and regulation of peroxiredoxins." Trends Biochem Sci 28(1): 32-40.
Yan, L. Z. and P. E. Dawson (2001). "Design and Synthesis of a Protein Catenane." Angew Chem Int Ed Engl 40(19): 3625-3627.
| a) | b) |
 |
 |