Java Based Developments for Protein Crystallography
 John W.
Campbell, November 2005
John W.
Campbell, November 2005
This article briefly describes a library of PX related Java
classes and two Java programs based on these which have similar functionality
to the CCP4 programs ROTGEN and IPDISP. An associated documentation preparation
program is also available.

The Java Development Library (JDL) is a set of Java classes written for
developing Java based software for Protein Crystallography. It contains four
main sections of classes as follows:
-
JdlLib
This is a set of general purpose classes which are used in the implementation
of the other sections of the library. These include classes and methods for
handling strings, files, data conversions and manipulating bits. In addition,
there are classes and methods for handling 3 element vectors, 3x3 matrices and
4x4 matrices and a class which carries out general non-linear least squares
refinements using the Levenberg-Marquardt algorithm.
-
JdlView

JdlView is a package for use in developing windows based programs based on an
older X-windows based toolkit
XDL_VIEW. The JdlView classes
basically fall into two categories. These are the general purpose JdlView
classes which are written without any particular application in mind (e.g the
menu area class or the I/O window class) and those which are written with a
particular application or group of applications in mind, particularly for
Protein Crystallography. They are primarily intended to be used in programs
which use the JdlView management class which forms part of the package but they
can also be used independently of that if required to fit within some
alternative programming context.
-
JdlPX

This is a set of classes specifically written for use in protein
crystallography. It includes classes for holding various sets of
crystallographic data, symmetry handling, reading and manipulating diffraction
images. There are classes and methods for predicting Rotation, Laue and Unique
reflection data and classes for storing and manipulating the predicted
reflection lists.
There is also a class which implements a
Diffraction Data Module (DDM) 
-
JdlKDM
This is a set of classes for defining and handling Keyworded parameter files
and their associated data.
Detailed
documentation of the JDL classes may be found
on the CCP4 website.

PXSim is a program for carrying out simulations of Protein Crystallographic
diffraction images and analysing the unique data coverage for one or more
crystal settings and series of diffraction images. It is intended primarily as
a teaching tool to enable the user to investigate, in some depth, the nature of
X-ray diffraction patterns and the amount of data which may be collected using
various settings or methods. The program may be used for simulating diffraction
patterns for the Rotation, Weissenberg and Laue methods of data collection and
analysing the extent of data which may be measured using those methods. The
predictions are based on the set of parameters defined in a
Diffraction Data Module (DDM).
The use of Java was chosen so that the program could be made available for use
across the internet as well as being in a form suitable for porting to any Java
supporting platform.
The basic screen layout is shown below:
Figure 1: Main
Screen of PXSim
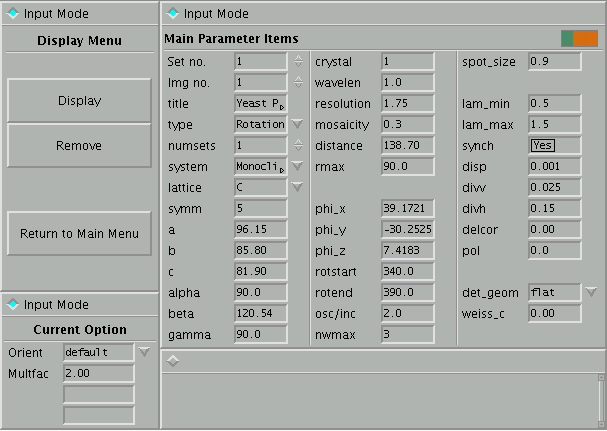
The main options of the programs are as follows:
-
Rotation/Weissenberg/Laue Simulations
Two basic types of simulations are available, standard colour simulations and
interactive colour simulations. In each case a window is created with a display
area for the simulation, an area to list details of a selected reflection, a
control panel and a button used for requesting printer or file (JPEG or PNG)
output of the simulation. The standard colour simulations show the show the
patterns with the spots colour coded in a number of different ways. The
interactive simulations have a slider which allow the user to investigate the
effects of changing various parameters such as the oscillation range and
mosaicity (or Lambda-min, dmin for Laue); spot labelling is also available and
it is possible to search for reflections by their indices.
An example of an 'interactive' simulation is shown below:
Figure 2: Example
of an Interactive Rotation Simulation
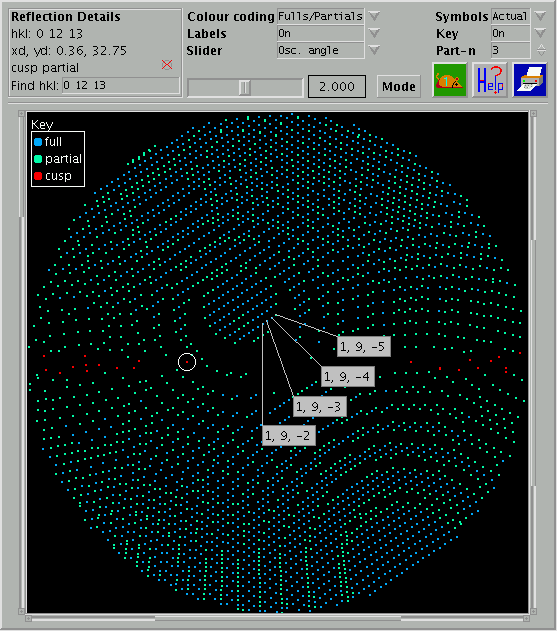
-
Unique Data Coverage
This option enables the prediction of the reflections which would be recorded
for the defined crystal sets and to analyse the data coverage in terms of the
unique data for the space group, cell and resolution. The analysis may be done
for either the current crystal set or for all crystal sets within the data set.
The results may be presented in the form or histograms describing the data
coverage, in terms of a pictorial representation of the reciprocal lattice
sections or as a three dimensional (rotatable) view of the reciprocal lattice.
As an example, I have chosen one of the three dimensional views as this is an
option which has been newly developed for this program.
Figure 3: Unique
Coverage - 3D Plot
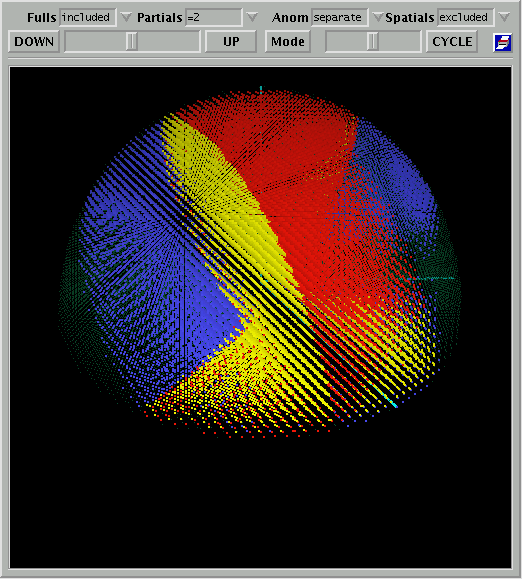
The program is available in
applet form on the CCP4 web
site.
Detailed documentation is also available
there.

This program incorporates most of the functionality of the program IPDISP. An
example is shown below:
Figure 4: PXImage
program - main screen
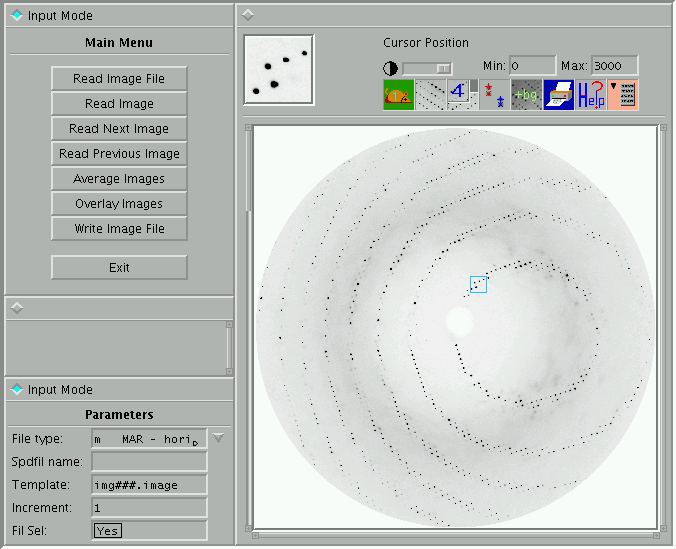
Some of the options for measuring the image are available via a drop down menu
and a dialogue window as shown below:
Figure 5: PXImage
program - measurement options dialogue
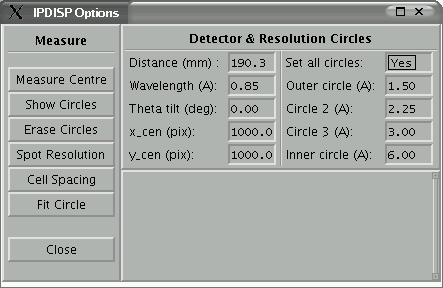
This program is still under development and is not yet available via the CCP4
web site though there is an
image display applet based on
the image JdlPXImage class which contains many of the available functions.

This program is used to prepare documentation files from in a 'pre-xhtml'
format or by extracting documentation sections from Java code source files. The
output files will normally be in XHTML format. Documentation may be prepared
from a single file or a set of linked document files. All internal links (and
an optional contents page) are generated automatically by the program. This
program has been used to prepare the documentation for the Java Development
Library, the Diffraction Data Module and the PXSim program.
 John W.
Campbell, November 2005
John W.
Campbell, November 2005 John W.
Campbell, November 2005
John W.
Campbell, November 2005




