HAPPy progress
Dan Rolfe, Charles Ballard, Maria Turkenburg*,
Eleanor Dodson*, Paul Emsley*
Daresbury Laboratory, Kekwick Lane, Warrington WA4
4AD
*
University
of
York
, York Y010 5YW
Work on the new experimental
phasing system, HAPPy (Heavy Atom Phasing in Python), discussed in the previous
newsletter, is progressing well. We are testing several packages for inclusion
as phasing modules including MLPHARE and PHASER. HAPPy can now take a SAD
dataset from the post data processing stage to a refined map with no
interaction from the user after providing the initial problem description. The
current priority is determining the optimal parameters for the major components
and systematic testing and analysis of results to improve the quality of
solutions obtained.
There has also been work on the
history and data tracking for the next generation CCP4i database and the
development of an experimental phasing datamodel for the database being
developed by CCP4 in the BioXHIT framework.
|
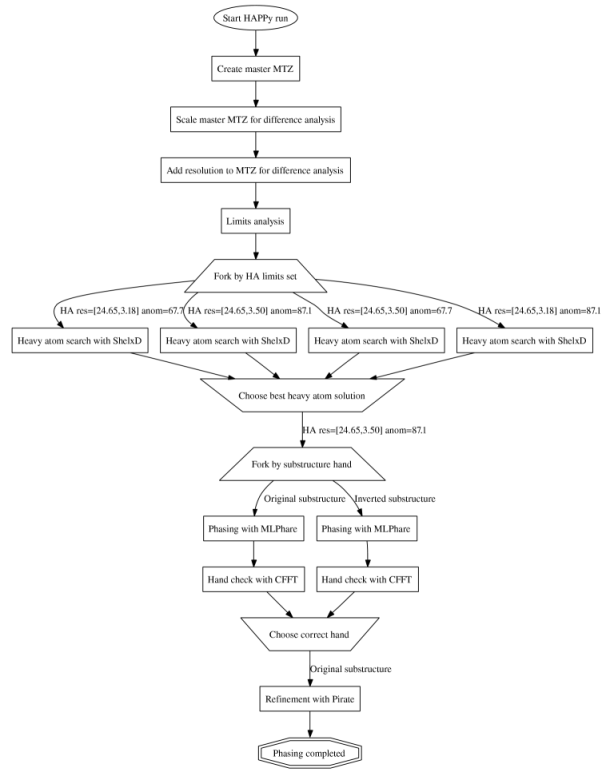
The diagram on the right, generated automatically
by the HAPPy tracking database, shows the flow of a simple example run. Note
that this shows what happened for this particular run; the procedure can become
more complicated depending on the dataset and options selected. In this
example, HAPPy first ran SHELXD with various sets of resolution and anomalous
difference limits, then chose the best resulting heavy atom solution for
phasing with MLPHARE. After determining the correct hand for the heavy atom
substructure by looking at variance maps of the MLPHARE output, the final map
and phases were refined using PIRATE. The figure below shows a section of the
final map in COOT.
|
|
|
 |

