![]()
using 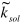 and
and 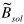 values defined in the input file;
values defined in the input file;
Guido Capitani1
and Andrei Fokine2
1Dept. of Biochemistry, University
of Zürich, Winterthurerstrasse 190,
CH-8057 Zürich, Switzerland
1LCM3B, UMR 7036 CNRS, Faculté des
Sciences, Université Henry
Poincaré, Nancy I,
54506 Vandoeuvre-lés-Nancy, France
Email: capitani@bioc.unizh.ch
Low-resolution reflections, being less sensitive to model imperfections, are known to be very useful (Urzhumtsev & Podjarny, 1995a, Fokine & Urzhumtsev, 2002a) for solving the translation problem in molecular replacement (Rossmann, 1972). In protein crystallography, traditional molecular replacement protocols, however, exclude reflections of resolution lower than 10 or 15 Å, because they are strongly influenced by the contribution of the bulk solvent in the crystals. As a consequence, at low resolution, comparison of structure factors calculated from the atomic model with experimental values is not reliable. In order to obviate this problem, a bulk solvent correction has to be introduced in the translation search procedure. Two different bulk solvent correction approaches are available (Kostrewa, 1997), the exponential scaling model (Moews & Kretsinger, 1975) and the mask model (Phillips, 1980; Jiang & Brünger, 1994), -both originally developed for macromolecular refinement purposes-. The exponential scaling model, based on Babinet's principle, is of simple implementation and is now applied to calculate a bulk solvent correction in the molecular replacement programs MOLREP (Vagin & Teplyakov, 1997), QS (Glykos & Kokkinidis, 2000) and BEAST (Read, 2001). The assumptions this approach is based on are, however, only true at resolutions below ~ 15 Å (Urzhumtsev & Podjarny, 1995b) and its performance was shown to be inferior to that of the mask model (Kostrewa, 1997). This latter method involves explicit calculation of an envelope around the protein model, creating a 'solvent mask'. The solvent region delimited by this solvent mask is then filled with bulk solvent electron density, and structure factors for this density are calculated and vectorially added to those derived from the protein model. Fokine and Urzhumtsev (2001) suggested a way to employ the mask method to calculate accurate bulk solvent correction for fast translation searches in molecular replacement. The corresponding program BULK (Fokine, Capitani, Grütter & Urzhumtsev, 2002) can be used with the standalone version of AMoRe (Navaza, 1994; Navaza & Vernoslova, 1995), with the CCP4 version of AMoRe (CCP4, 1994) and with CNS (Brünger et al., 1998).
A typical molecular replacement run with AMoRe involves three main steps: the calculation of a structure factor (SF) table from the search model (often referred to as 'tabling step'), the cross-rotation function search (' roting step') and the fast translation search (' traing step'). The table below summarizes those steps and their input and output files:
| INPUT | STEP | OUTPUT |
| Search model (PDB file) | Tabling | SF table (' search tab') 'tabbed model'* Tabling step log file |
| Experimental amplitudes SF table (' search. tab') |
Roting | List of cross-rotation function peaks |
| Experimental amplitudes SF table (' search tab') List of cross-rotation function peaks |
Traing | List of translation function peaks |
The program BULK introduces another step (' bulking') in this procedure and calculates a bulk-solvent corrected structure factor table to be used in the 'traing' step. The scheme is then modified as follows:
| INPUT | STEP | OUTPUT |
| Search model (PDB file) | Tabling | SF table (' search tab') 'tabbed model' Tabling step log file |
| Experimental amplitudes SF table (' search. tab') |
Roting | List of cross-rotation function peaks |
| SF table (' search. tab') Search model Tabling step log file |
Bulking | Corrected SF table (search tabs) |
| Experimental amplitudes Corrected SF table (search tabs) List of cross-rotation function peaks |
Traing | List of translation function peaks |
The 'bulking' procedure encompasses various computational tasks, which are to be carried out after the AMoRe 'tabling' step, where the search model is placed in a large rectangular box (the box dimensions are determined automatically) with its centre of mass at the origin and its principal inertia axes parallel to the box axes. A model rotated and translated in this way can be referred to as a 'tabbed' model. Then, structure factors FmP1( h) from this model are calculated by AMoRe. 'BULK' goes then through the following steps:
![]()
using 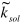 and
and 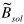 values defined in the input file;
values defined in the input file;
The theoretical ground for the above calculations is discussed in
(Fokine,
Capitani, Grütter & Urzhumtsev, 2002). Here we just point out that
the
procedure uses constant solvent scaling and B-factor parameters
(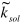 ,
, 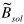 ) as defined by an input file. Mean values for these parameters (0.35
e/Å3 and
46 Å2 , respectively) were derived by Fokine & Urzhumtsev
(2002b) through a
statistical analysis of well-refined structures deposited in the Protein
Data
Bank (Bernstein et al., 1977). If for a certain crystal the
buffer
density differs markedly from standard values, the user can accordingly
change
the default
) as defined by an input file. Mean values for these parameters (0.35
e/Å3 and
46 Å2 , respectively) were derived by Fokine & Urzhumtsev
(2002b) through a
statistical analysis of well-refined structures deposited in the Protein
Data
Bank (Bernstein et al., 1977). If for a certain crystal the
buffer
density differs markedly from standard values, the user can accordingly
change
the default 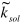 and
and 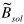 input parameters. Another point worth mentioning is that 'BULK' can be
used only
in cases when one entity in the asymmetric unit is searched for (this
can be
also a tetramer or an octamer, provided that a corresponding tetrameric
or
octameric search model, respectively, is available). On the contrary, if
the
molecular replacement problem involves, for instance, locating two
independent
monomers, the current approach cannot be used because it would produce
unreliable structure factors (Fokine, Capitani, Grütter &
Urzhumtsev, 2002).
input parameters. Another point worth mentioning is that 'BULK' can be
used only
in cases when one entity in the asymmetric unit is searched for (this
can be
also a tetramer or an octamer, provided that a corresponding tetrameric
or
octameric search model, respectively, is available). On the contrary, if
the
molecular replacement problem involves, for instance, locating two
independent
monomers, the current approach cannot be used because it would produce
unreliable structure factors (Fokine, Capitani, Grütter &
Urzhumtsev, 2002).
To install and run BULK one first has to copy the 'bulk' directory (as extracted from the BULK distribution tar file) into the AMoRe working directory, then to compile it with the command:
./bulk/make_bulkThis creates two executables, prep_bulk and bulking.
The only input file needed, (an example is provided in the distribution), is prep_bulk.inp, with the following contents:
| search.pdb | name of the file with model coordinates (the same as used for the 'tabling'step) |
| search.tab | name of the file of structure factors from the AMoRe'tabling' step |
| tab.log | name of the log file of the AMoRe 'tabling' step |
| search.tabs | name of the file with bulk-solvent-corrected structure factors (this file is created by the 'bulking' procedure) |
| bulking.inp | name of an intermediate control file to be input to 'bulking';
this file is created by running 'prep_bulk' |
| 0.35 | value of 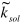 (scaling parameter for solvent electron density, in
e/Å3 )
(scaling parameter for solvent electron density, in
e/Å3 ) |
| 50.0 | value of 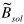 (temperature factor for solvent, in Å2
)
(temperature factor for solvent, in Å2
) |
| 1.0 | value of the solvent radius used for the solvent mask calculation |
To run the procedure, one issues the two following shell commands:
prep_bulk < prep_bulk.inp
bulking < bulking.inp
'prep_bulk' creates the file 'bulking.inp', containing the
information
needed by 'bulking' to calculate the actual bulk-solvent correction. The
output
file 'search.tabs', created by 'bulking', contains the
bulk-solvent-corrected
structure factors and can be used for the translation search instead of
'search.tab'. Importantly, the corrected structure factor file should
not be
used in the 'roting' step, since low-resolution reflections contribute
rather
negatively to the rotation function search results.
A conventional rotation search was then carried out in the range 8-4 Å and the top 30 rotation function solutions were listed. BULK was then used to calculate a corrected structure factor table. Translation tests were performed in the ranges 10-4 Å and 15-4 Å, both with and without bulk solvent correction. The top 30 solutions in each case, sorted by correlation coefficient, are reported below (α,β and γ are the eulerian rotation angles for each solution, Tx, Ty, Tz the fractional translations, cc, Rf and cc-I the amplitude-based correlation coefficient, the R-factor and the intensity-based correlation coefficient, respectively):
| α | β | γ | Tx | Ty | Tz | cc | Rf | cc-I | ||
| SOLUTIONTF1 | 1 | 139.59 | 56.87 | 225.89 | 0.1691 | 0.4390 | 0.2361 | 9.2 | 55.6 | 9.5 |
| SOLUTIONTF1 | 1 | 134.00 | 59.87 | 50.32 | 0.1338 | 0.4407 | 0.4377 | 6.0 | 57.2 | 8.3 |
| SOLUTIONTF1 | 1 | 72.06 | 41.69 | 133.77 | 0.3280 | 0.2580 | 0.0577 | 5.4 | 57.4 | 6.2 |
| SOLUTIONTF1 | 1 | 145.89 | 45.72 | 44.62 | 0.4242 | 0.1225 | 0.0964 | 5.2 | 57.3 | 5.1 |
| SOLUTIONTF1 | 1 | 88.83 | 90.00 | 60.70 | 0.2029 | 0.1829 | 0.1960 | 5.2 | 58.8 | 5.4 |
| SOLUTIONTF1 | 1 | 68.57 | 36.76 | 316.05 | 0.1218 | 0.4297 | 0.3087 | 5.2 | 57.6 | 4.5 |
| SOLUTIONTF1 | 1 | 60.70 | 42.44 | 244.79 | 0.3353 | 0.3195 | 0.1074 | 4.6 | 56.8 | 5.0 |
| SOLUTIONTF1 | 1 | 111.50 | 36.91 | 56.00 | 0.2603 | 0.3188 | 0.1500 | 4.5 | 56.9 | 5.1 |
| SOLUTIONTF1 | 1 | 113.50 | 46.00 | 58.30 | 0.2170 | 0.0728 | 0.2099 | 4.2 | 57.7 | 5.1 |
| SOLUTIONTF1 | 1 | 86.00 | 90.00 | 18.47 | 0.4114 | 0.9958 | 0.0000 | 4.2 | 61.3 | 3.9 |
| SOLUTIONTF1 | 1 | 95.24 | 90.00 | 69.66 | 0.4683 | 0.2917 | 0.1845 | 4.0 | 57.8 | 3.6 |
| SOLUTIONTF1 | 1 | 43.81 | 38.01 | 48.64 | 0.2352 | 0.2299 | 0.0202 | 4.0 | 56.9 | 3.0 |
| SOLUTIONTF1 | 1 | 116.65 | 35.64 | 233.53 | 0.0059 | 0.0018 | 0.4482 | 3.9 | 57.3 | 5.4 |
| SOLUTIONTF1 | 1 | 87.08 | 26.20 | 306.42 | 0.3940 | 0.3887 | 0.1388 | 3.9 | 57.8 | 4.6 |
| SOLUTIONTF1 | 1 | 56.96 | 81.80 | 194.07 | 0.1389 | 0.0040 | 0.1650 | 3.9 | 57.4 | 3.3 |
| SOLUTIONTF1 | 1 | 54.21 | 51.00 | 39.16 | 0.3014 | 0.3577 | 0.3644 | 3.8 | 57.4 | 2.7 |
| SOLUTIONTF1 | 1 | 15.39 | 70.84 | 128.24 | 0.0188 | 0.0999 | 0.4365 | 3.7 | 58.2 | 3.5 |
| SOLUTIONTF1 | 1 | 158.40 | 86.17 | 268.98 | 0.4258 | 0.3079 | 0.0769 | 3.5 | 57.8 | 2.7 |
| SOLUTIONTF1 | 1 | 91.00 | 29.96 | 121.50 | 0.3952 | 0.3952 | 0.1324 | 3.5 | 57.7 | 4.5 |
| SOLUTIONTF1 | 1 | 83.68 | 80.82 | 311.33 | 0.1649 | 0.1792 | 0.0221 | 3.5 | 57.6 | 4.9 |
| SOLUTIONTF1 | 1 | 76.30 | 36.00 | 311.71 | 0.3069 | 0.2979 | 0.0116 | 3.5 | 58.0 | 4.1 |
| SOLUTIONTF1 | 1 | 57.79 | 87.04 | 284.00 | 0.3811 | 0.2135 | 0.4233 | 3.5 | 57.6 | 2.7 |
| SOLUTIONTF1 | 1 | 94.96 | 70.55 | 251.19 | 0.1450 | 0.1805 | 0.2657 | 3.4 | 57.5 | 3.6 |
| SOLUTIONTF1 | 1 | 80.54 | 34.85 | 309.13 | 0.0546 | 0.0800 | 0.2310 | 3.4 | 58.0 | 4.0 |
| SOLUTIONTF1 | 1 | 51.00 | 35.70 | 221.50 | 0.2660 | 0.1106 | 0.0859 | 3.2 | 57.5 | 2.8 |
| SOLUTIONTF1 | 1 | 92.23 | 90.00 | 197.89 | 0.3074 | 0.0060 | 0.0000 | 3.1 | 63.3 | 3.5 |
| SOLUTIONTF1 | 1 | 11.86 | 64.00 | 129.40 | 0.3105 | 0.4471 | 0.4815 | 3.1 | 57.2 | 3.7 |
| SOLUTIONTF1 | 1 | 151.00 | 41.61 | 314.00 | 0.3379 | 0.4513 | 0.2173 | 2.9 | 58.2 | 2.6 |
| SOLUTIONTF1 | 1 | 43.60 | 81.50 | 13.16 | 0.1041 | 0.3664 | 0.3687 | 2.8 | 57.7 | 3.3 |
| SOLUTIONTF1 | 1 | 86.40 | 86.41 | 247.14 | 0.2704 | 0.4661 | 0.2848 | 2.5 | 57.5 | 2.7 |
This first case represents a 'classical' resolution range for a translation search run. Sorting by correlation coefficient identifies one slightly emerging solution but the signal is weak. Further analysis, including comparison with the newly solved structure, shows that the top-ranking solution (in blue, with cyan background) is correct, whereas the second is wrong.
| α | β | γ | Tx | Ty | Tz | cc | Rf | cc-I | ||
| SOLUTIONTF1 | 1 | 134.00 | 59.87 | 50.32 | 0.1824 | 0.4440 | 0.2365 | 11.2 | 54.9 | 13.4 |
| SOLUTIONTF1 | 1 | 145.89 | 45.72 | 44.62 | 0.1943 | 0.4757 | 0.2176 | 9.9 | 54.5 | 9.7 |
| SOLUTIONTF1 | 1 | 72.06 | 41.69 | 133.77 | 0.2079 | 0.1879 | 0.4616 | 9.1 | 55.3 | 9.2 |
| SOLUTIONTF1 | 1 | 113.50 | 46.00 | 58.30 | 0.2201 | 0.0987 | 0.2040 | 8.5 | 55.4 | 9.8 |
| SOLUTIONTF1 | 1 | 111.50 | 36.91 | 56.00 | 0.4681 | 0.3272 | 0.3476 | 8.3 | 55.0 | 8.1 |
| SOLUTIONTF1 | 1 | 88.83 | 90.00 | 60.70 | 0.2028 | 0.1860 | 0.1941 | 8.3 | 57.2 | 8.1 |
| SOLUTIONTF1 | 1 | 68.57 | 36.76 | 316.05 | 0.2626 | 0.0711 | 0.4542 | 8.1 | 55.3 | 8.5 |
| SOLUTIONTF1 | 1 | 57.79 | 87.04 | 284.00 | 0.3830 | 0.2146 | 0.4251 | 7.8 | 55.3 | 6.9 |
| SOLUTIONTF1 | 1 | 15.39 | 70.84 | 128.24 | 0.3441 | 0.9989 | 0.4286 | 7.8 | 55.4 | 7.5 |
| SOLUTIONTF1 | 1 | 76.30 | 36.00 | 311.71 | 0.3643 | 0.4896 | 0.2877 | 7.7 | 55.5 | 8.8 |
| SOLUTIONTF1 | 1 | 158.40 | 86.17 | 268.98 | 0.1451 | 0.4607 | 0.0658 | 7.5 | 55.5 | 7.7 |
| SOLUTIONTF1 | 1 | 95.24 | 90.00 | 69.66 | 0.3852 | 0.0560 | 0.3756 | 7.4 | 55.2 | 7.0 |
| SOLUTIONTF1 | 1 | 116.65 | 35.64 | 233.53 | 0.0096 | 0.4709 | 0.4455 | 7.3 | 55.1 | 8.0 |
| SOLUTIONTF1 | 1 | 80.54 | 34.85 | 309.13 | 0.0566 | 0.0790 | 0.2314 | 7.3 | 55.9 | 8.8 |
| SOLUTIONTF1 | 1 | 56.96 | 81.80 | 194.07 | 0.4703 | 0.3603 | 0.1054 | 7.3 | 55.1 | 6.9 |
| SOLUTIONTF1 | 1 |
94.96 | 70.55 | 251.19 | 0.1271 | 0.4514 | 0.3930 | 7.1 | 55.6 | 6.8 |
| SOLUTIONTF1 | 1 |
60.70 | 42.44 | 244.79 | 0.4287 | 0.3201 | 0.0630 | 7.1 | 54.6 | 7.4 |
| SOLUTIONTF1 | 1 |
54.21 | 51.00 | 39.16 | 0.3420 | 0.2334 | 0.0730 | 7.1 | 55.5 | 7.3 |
| SOLUTIONTF1 | 1 |
87.08 | 26.20 | 306.42 | 0.2921 | 0.0411 | 0.4443 | 6.9 | 55.5 | 7.4 |
| SOLUTIONTF1 | 1 |
83.68 | 80.82 | 311.33 | 0.2528 | 0.4874 | 0.1895 | 6.9 | 56.3 | 9.3 |
| SOLUTIONTF1 | 1 |
11.86 | 64.00 | 129.40 | 0.3803 | 0.4433 | 0.1825 | 6.9 | 55.7 | 8.0 |
| SOLUTIONTF1 | 1 |
86.00 | 90.00 | 18.47 | 0.4117 | 0.9946 | 0.0000 | 6.8 | 59.9 | 5.9 |
| SOLUTIONTF1 | 1 |
43.81 | 38.01 | 48.64 | 0.2616 | 0.0488 | 0.3946 | 6.7 | 55.4 | 6.0 |
| SOLUTIONTF1 | 1 |
51.00 | 35.70 | 221.50 | 0.4401 | 0.1680 | 0.0470 | 6.4 | 55.4 | 6.2 |
| SOLUTIONTF1 | 1 |
92.23 | 90.00 | 197.89 | 0.4567 | 0.0409 | 0.2660 | 6.3 | 58.8 | 5.5 |
| SOLUTIONTF1 | 1 |
86.40 | 86.41 | 247.14 | 0.1486 | 0.4503 | 0.2280 | 6.3 | 56.2 | 7.4 |
| SOLUTIONTF1 | 1 |
43.60 | 81.50 | 13.16 | 0.1480 | 0.4897 | 0.2492 | 6.3 | 56.3 | 6.4 |
| SOLUTIONTF1 | 1 |
91.00 | 29.96 | 121.50 | 0.2939 | 0.2384 | 0.4492 | 6.1 | 55.8 | 6.3 |
| SOLUTIONTF1 | 1 |
151.00 | 41.61 | 314.00 | 0.0900 | 0.2623 | 0.3265 | 5.7 | 56.2 | 6.2 |
In this run the resolution range is the same as before, but the first solution (also same as before) now exhibits a much higher correlation coefficient. By comparing the first to the second ranking solution (both in blue), it appears that they are both correct and equivalent (same translations and rotations, except for a ~ 180° difference in γ, which is readily explained by the two-fold symmetry of the search model). Interestingly, also in run A) a similar situation is observed, but the value for Tz in the second solution differs from that of the first and turns out to be wrong by further analysis. Comparison of runs A) and B) shows then that using the bulk solvent correction was instrumental for obtaining correct translation parameters for the second solution.
| α | β | γ | Tx | Ty | Tz | cc | Rf | cc-I | ||
| SOLUTIONTF1 | 1 | 139.59 | 56.87 | 225.89 | 0.1716 | 0.4378 | 0.2365 | 16.4 | 53.5 | 16.8 |
| SOLUTIONTF1 | 1 | 134.00 | 59.87 | 50.32 | 0.1846 | 0.4415 | 0.2346 | 12.7 | 54.8 | 14.2 |
| SOLUTIONTF1 | 1 | 145.89 | 45.72 | 44.62 | 0.1950 | 0.4740 | 0.2176 | 11.1 | 54.7 | 9.7 |
| SOLUTIONTF1 | 1 | 113.50 | 46.00 | 58.30 | 0.3360 | 0.3380 | 0.3054 | 9.0 | 55.9 | 9.7 |
| SOLUTIONTF1 | 1 | 68.57 | 36.76 | 316.05 | 0.3924 | 0.4734 | 0.2250 | 8.9 | 56.4 | 9.0 |
| SOLUTIONTF1 | 1 | 111.50 | 36.91 | 56.00 | 0.0206 | 0.0148 | 0.4481 | 8.8 | 55.6 | 8.9 |
| SOLUTIONTF1 | 1 | 88.83 | 90.00 | 60.70 | 0.2027 | 0.1848 | 0.1937 | 8.8 | 57.6 | 8.3 |
| SOLUTIONTF1 | 1 | 72.06 | 41.69 | 133.77 | 0.1529 | 0.3820 | 0.4211 | 8.8 | 55.9 | 9.9 |
| SOLUTIONTF1 | 1 | 158.40 | 86.17 | 268.98 | 0.4053 | 0.4080 | 0.0285 | 8.4 | 55.6 | 6.8 |
| SOLUTIONTF1 | 1 | 60.70 | 42.44 | 244.79 | 0.3510 | 0.2223 | 0.1524 | 7.9 | 55.4 | 6.7 |
| SOLUTIONTF1 | 1 | 76.30 | 36.00 | 311.71 | 0.3651 | 0.4889 | 0.2886 | 7.8 | 56.2 | 7.4 |
| SOLUTIONTF1 | 1 | 116.65 | 35.64 | 233.53 | 0.0059 | 0.0035 | 0.4482 | 7.7 | 55.9 | 8.7 |
| SOLUTIONTF1 | 1 | 94.96 | 70.55 | 251.19 | 0.0268 | 0.2212 | 0.2433 | 7.6 | 55.8 | 6.5 |
| SOLUTIONTF1 | 1 | 83.68 | 80.82 | 311.33 | 0.4264 | 0.0023 | 0.3584 | 7.2 | 56.7 | 8.0 |
| SOLUTIONTF1 | 1 | 80.54 | 34.85 | 309.13 | 0.0563 | 0.0787 | 0.2313 | 7.2 | 56.4 | 7.3 |
| SOLUTIONTF1 | 1 | 15.39 | 70.84 | 128.24 | 0.2261 | 0.1199 | 0.4634 | 7.2 | 56.2 | 6.8 |
| SOLUTIONTF1 | 1 | 11.86 | 64.00 | 129.40 | 0.1529 | 0.1712 | 0.1759 | 7.0 | 55.5 | 6.6 |
| SOLUTIONTF1 | 1 | 43.81 | 38.01 | 48.64 | 0.4538 | 0.1625 | 0.0477 | 6.9 | 56.1 | 6.5 |
| SOLUTIONTF1 | 1 | 56.96 | 81.80 | 194.07 | 0.0170 | 0.3098 | 0.1236 | 6.7 | 55.9 | 5.8 |
| SOLUTIONTF1 | 1 | 43.60 | 81.50 | 13.16 | 0.1484 | 0.4888 | 0.2491 | 6.7 | 56.8 | 6.1 |
| SOLUTIONTF1 | 1 | 54.21 | 51.00 | 39.16 | 0.4464 | 0.3624 | 0.2688 | 6.6 | 55.8 | 4.6 |
| SOLUTIONTF1 | 1 | 95.24 | 90.00 | 69.66 | 0.3267 | 0.4105 | 0.0458 | 6.5 | 56.4 | 6.6 |
| SOLUTIONTF1 | 1 | 87.08 | 26.20 | 306.42 | 0.2932 | 0.0408 | 0.4435 | 6.5 | 56.2 | 6.5 |
| SOLUTIONTF1 | 1 | 86.00 | 90.00 | 18.47 | 0.4130 | 0.9969 | 0.2583 | 6.5 | 58.3 | 5.7 |
| SOLUTIONTF1 | 1 | 57.79 | 87.04 | 284.00 | 0.2481 | 0.2693 | 0.4858 | 6.3 | 56.0 | 6.3 |
| SOLUTIONTF1 | 1 | 51.00 | 35.70 | 221.50 | 0.4421 | 0.1697 | 0.0471 | 6.3 | 55.9 | 5.9 |
| SOLUTIONTF1 | 1 | 86.40 | 86.41 | 247.14 | 0.1754 | 0.2091 | 0.4515 | 6.1 | 56.4 | 6.6 |
| SOLUTIONTF1 | 1 | 151.00 | 41.61 | 314.00 | 0.0314 | 0.2624 | 0.1190 | 5.8 | 56.4 | 5.7 |
| SOLUTIONTF1 | 1 | 92.23 | 90.00 | 197.89 | 0.3987 | 1.0000 | 0.2551 | 5.8 | 59.5 | 4.8 |
| SOLUTIONTF1 | 1 | 91.00 | 29.96 | 121.50 | 0.0376 | 0.3296 | 0.3309 | 5.7 | 56.6 | 6.7 |
In run C) the full available resolution range was used with bulk solvent correction, the results are similar to those of run B), but with higher correlation coefficients (also cc-I) for the first-and second-ranking solution (both in blue).
| α | β | γ | Tx | Ty | Tz | cc | Rf | cc-I | ||
| SOLUTIONTF1 | 1 | 139.59 | 56.87 | 225.89 | 0.1678 | 0.4373 | 0.2365 | 9.7 | 58.3 | 8.3 |
| SOLUTIONTF1 | 1 | 134.00 | 59.87 | 50.32 | 0.1305 | 0.4409 | 0.4365 | 7.3 | 59.4 | 7.1 |
| SOLUTIONTF1 | 1 | 145.89 | 45.72 | 44.62 | 0.2279 | 0.2650 | 0.3167 | 6.2 | 59.8 | 5.7 |
| SOLUTIONTF1 | 1 | 111.50 | 36.91 | 56.00 | 0.3655 | 0.2451 | 0.0275 | 6.1 | 59.9 | 6.1 |
| SOLUTIONTF1 | 1 | 113.50 | 46.00 | 58.30 | 0.1324 | 0.2200 | 0.0288 | 5.7 | 59.7 | 6.0 |
| SOLUTIONTF1 | 1 | 68.57 | 36.76 | 316.05 | 0.4704 | 0.4331 | 0.3752 | 5.6 | 60.0 | 5.1 |
| SOLUTIONTF1 | 1 | 94.96 | 70.55 | 251.19 | 0.0260 | 0.2183 | 0.4526 | 5.3 | 59.9 | 4.7 |
| SOLUTIONTF1 | 1 | 72.06 | 41.69 | 133.77 | 0.4068 | 0.4374 | 0.1118 | 5.0 | 59.9 | 5.5 |
| SOLUTIONTF1 | 1 | 158.40 | 86.17 | 268.98 | 0.4706 | 0.1000 | 0.3846 | 4.9 | 60.3 | 4.8 |
| SOLUTIONTF1 | 1 | 88.83 | 90.00 | 60.70 | 0.3382 | 0.3900 | 0.1923 | 4.8 | 62.2 | 4.1 |
| SOLUTIONTF1 | 1 | 60.70 | 42.44 | 244.79 | 0.0529 | 0.2115 | 0.3682 | 4.7 | 60.2 | 5.4 |
| SOLUTIONTF1 | 1 | 56.96 | 81.80 | 194.07 | 0.1636 | 0.0649 | 0.4190 | 4.6 | 59.4 | 3.9 |
| SOLUTIONTF1 | 1 | 87.08 | 26.20 | 306.42 | 0.2059 | 0.2900 | 0.2788 | 4.3 | 60.4 | 3.5 |
| SOLUTIONTF1 | 1 | 83.68 | 80.82 | 311.33 | 0.4454 | 0.1092 | 0.0475 | 4.3 | 60.6 | 4.3 |
| SOLUTIONTF1 | 1 | 43.81 | 38.01 | 48.64 | 0.4706 | 0.2500 | 0.2596 | 4.3 | 59.5 | 3.1 |
| SOLUTIONTF1 | 1 | 80.54 | 34.85 | 309.13 | 0.2072 | 0.2827 | 0.3633 | 4.0 | 60.9 | 3.8 |
| SOLUTIONTF1 | 1 | 76.30 | 36.00 | 311.71 | 0.0452 | 0.1494 | 0.0303 | 4.0 | 60.7 | 5.2 |
| SOLUTIONTF1 | 1 | 116.65 | 35.64 | 233.53 | 0.2206 | 0.2600 | 0.1058 | 3.9 | 60.3 | 4.5 |
| SOLUTIONTF1 | 1 | 43.60 | 81.50 | 13.16 | 0.1034 | 0.3654 | 0.3704 | 3.8 | 60.0 | 4.0 |
| SOLUTIONTF1 | 1 | 11.86 | 64.00 | 129.40 | 0.2546 | 0.1097 | 0.2098 | 3.7 | 60.4 | 3.2 |
| SOLUTIONTF1 | 1 | 95.24 | 90.00 | 69.66 | 0.3196 | 0.2313 | 0.4455 | 3.6 | 60.6 | 2.9 |
| SOLUTIONTF1 | 1 | 92.23 | 90.00 | 197.89 | 0.0335 | 0.3088 | 0.2327 | 3.6 | 63.4 | 3.0 |
| SOLUTIONTF1 | 1 | 15.39 | 70.84 | 128.24 | 0.0600 | 0.0896 | 0.1717 | 3.6 | 60.8 | 3.3 |
| SOLUTIONTF1 | 1 | 91.00 | 29.96 | 121.50 | 0.2794 | 0.3400 | 0.4038 | 3.5 | 60.8 | 3.6 |
| SOLUTIONTF1 | 1 | 86.40 | 86.41 | 247.14 | 0.2928 | 0.2094 | 0.4518 | 3.4 | 60.6 | 2.8 |
| SOLUTIONTF1 | 1 | 51.00 | 35.70 | 221.50 | 0.3235 | 0.2900 | 0.3269 | 3.4 | 60.5 | 2.9 |
| SOLUTIONTF1 | 1 | 86.00 | 90.00 | 18.47 | 0.1664 | 0.3998 | 0.0288 | 3.2 | 62.2 | 3.0 |
| SOLUTIONTF1 | 1 | 57.79 | 87.04 | 284.00 | 0.1324 | 0.2700 | 0.4904 | 3.1 | 60.2 | 2.8 |
| SOLUTIONTF1 | 1 | 54.21 | 51.00 | 39.16 | 0.4559 | 0.3600 | 0.3654 | 2.9 | 60.3 | 1.7 |
| SOLUTIONTF1 | 1 | 151.00 | 41.61 | 314.00 | 0.0987 | 0.3222 | 0.0750 | 2.5 | 60.2 | 3.3 |
Run D) shows the importance of applying the bulk solvent correction when using low-resolution reflections down to 15 Å: the results are in fact much poorer compared to those of run C) and similar to those of run A). Also in this case the Tz translation is wrong for the second-ranking solution (in blue).
BULK has been tested with CCP4 up to version 4.2.1. It is at the moment distributed upon request by the authors at the following addresses: fokine@lcm3b.uhp-nancy.fr, sacha@lcm3b.uhp-nancy.fr and capitani@bioc.unizh.ch. Future developments will involve the integration of BULK into the CCP4 distribution.
The authors are grateful to Profs. Alexandre Urzhumtsev and Markus G. Grütter for support and encouragement. G. C. wishes to acknowledge the Baugarten Foundation, Zürich, as well as Andreas Schweizer and Christophe Briand. A. F. acknowledges Lorraine Regional administration, Pole "Intelligence Logiciels" CPER-Lorraine and GdR 2417 CNRS for the financial support.
Bernstein, F. C., Koetzle, T. F., Williams, G. J., Meyer, E. F. Jr., Brice, M. D., Rodgers, J. R., Kennard, O., Shimanouchi, T. & Tasumi, M. (1977). J. Mol. Biol., 112, 535-542.
Brünger, A.T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst., D54, 905-921.
Collaborative Computational Project, Number 4 (1994). Acta Cryst., D50, 760-763.
Fokine, A., Capitani, G., Grütter, M. G. & Urzhumtsev A. (2002). J. Appl. Cryst., submitted.
Fokine, A. & Urzhumtsev A. (2001). CCP4 Newsletter, 40.
Fokine, A. & Urzhumtsev A. (2002a). Acta Cryst., A58, 72-74.
Fokine, A. & Urzhumtsev, A. (2002b). Acta Cryst. D58, 1387-92.
Glykos, N. M. & Kokkinidis, M. (2000). Acta Cryst. D56, 1070-1072.
Jiang, J.-S. & Brünger, A. (1994). J. Mol. Biol., 243, 100-115.
Kostrewa, D. (1997). CCP4 Newsletter, 34, 9-22.
Moews, P. C. & Kretsinger, R. H. (1975). J. Mol. Biol., 91, 201-225.
Navaza, J. (1994). Acta Cryst. A50, 157-163.
Navaza, J. & Vernoslova, E. (1995). Acta Cryst., A51, 445-449.
Phillips, S. E. V. (1980). J. Mol. Biol., 142, 531-554.
Read, R. J. (1997). Acta Cryst. D57, 1373-1382.
Rossmann, M. G., ed. (1972). The Molecular Replacement Method, Gordon & Breach; New York, London, Paris.
Urzhumtsev, A. & Podjarny, A. D. (1995a). Acta Cryst., D51, 888-895.
Urzhumtsev, A. & Podjarny, A. D. (1995b). Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography, 31, 12-16.
Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst. 30, 1022-1025.