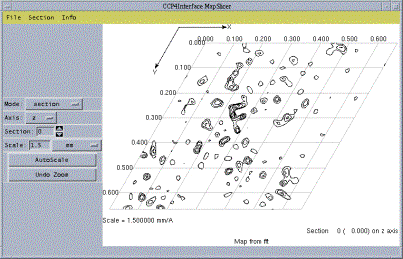
Figure 1: screenshot of MapSlicer
Peter Briggs, Pryank Patel, Alun Ashton, Charles Ballard, Liz Potterton*, Maria Turkenburg*, Martyn Winn
CCP4, Daresbury Laboratory, Warrington WA4 4AD, UK
*Structural Biology Laboratory, Department of Chemistry, University of York Y010 5YW, UK
CCP4i is the CCP4 graphical user interface. The last officially released version of the interface was 1.3.8, included as part of CCP4 4.2.1. This article outlines the major changes in 1.3.8 against the previous version of CCP4i, and looks ahead to some of the future developments planned for the next release and beyond.
Many of the new and updated features in the current version of CCP4i were previewed in the previous newsletter (issue 40 March 2002). As well as a number of relatively minor changes to fix bugs and consolidate earlier changes, there were a number of new and updated task interfaces, reflecting changes and additions to the suite in release 4.2.
These include interfaces for CCP4 programs ANISOANL, TLSANL and OASIS, as well as interfaces for the major new programs in 4.2: ACORN (ab initio procedure for the determination of protein structure at atomic resolution), BEAST (maximum-likelihood molecular replacement program), PROFESSS (determination of NCS operators from heavy atom substructure) and ROTAMER (comparison of atomic coordinates against Richardson's Penultimate Rotamer Library).
In addition, the GET_PROT interface (now renamed SAPHIRE) is a CCP4i-only application which allows the user to download and edit protein sequence files accessed either from the EBI or from a local file. An interface for WHAT_CHECK (the subset of protein verification tools from the WHAT IF program) is also included although the program itself is not yet distributed with the CCP4 suite. (See http://www.cmbi.kun.nl/whatif/ for more information on WHAT IF.)
Major changes have been made to the SCALA interface, to accommodate changes in the handling of datasets. A number of other minor changes have been made to existing interfaces in an attempt to improve ease of use, for example the Scalepack2mtz and Dtrek2mtz tasks have been combined into a single task interface to import scaled data. Also there has been some reorganisation of the tasks and modules menus (for example the addition of a ``Validation and Deposition'' module) to improve access to relevant tasks at various stages of the structure solution process.
MapSlicer offers interactive display of contoured 2D sections through CCP4-format density maps. MapSlicer has been significantly improved between CCP4 4.1 and 4.2 (including a substantial redesign of its own user interface) and is now built as a standard part of the default for many platforms. As well as allowing the user to flip between different sections and map axes, the program also allows the display of ``slabs'' of multiple sections, and the ability to go directly to Harker sections.
Although not strictly CCP4i, MapSlicer uses many components from the interface and so maintains a similiar ``look and feel''. Also, it is now possible for the user to set their preferences to make MapSlicer the default viewer for maps when accessed via the ``View Files from Job'' menu on the main CCP4i window.

Figure 1: screenshot of MapSlicer
The range of features offered by MapSlicer is still relatively modest, for example displays are still only black and white. A number of possible improvements are envisaged, including a built in peaksearch algorithm, and display of coordinates - for example heavy atom positions or peaks corresponding to calculated Harker vectors.
AstexViewer is a Java application written by Mike Hartshorn to display density maps and protein-ligand complexes. One way of using the viewer is to embed it as an applet in a webpage and then view it using your favourite browser. CCP4i now includes a task interface which will generate these pages automatically and launch a browser to view them.
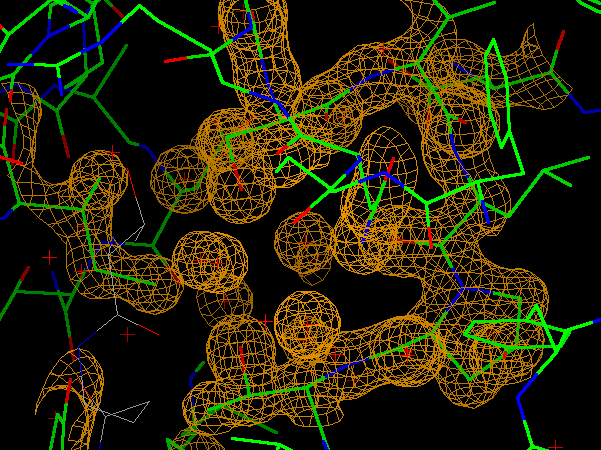
Figure 2: gif output from the AstexViewer, showing a protein structure inside a density map
The Task Installer utility for installing new task interfaces has been substantially upgraded, and aims to provide a robust mechanism for installing and tracking ``third-party'' interfaces - that is, interfaces provided for non-CCP4 software by the authors of that software. An example of this is the CCP4i interface for ARP/wARP, written by Tassos Perrakis and distributed with the latest version of the ARP/wARP suite (version 6.0 - see http://www.arp-warp.org for more details).
For users, the new utility offers options to install, review and uninstall these interfaces quickly and easily. New interfaces can also be installed either ``locally'' (so only the person installing the task can use it) or ``publically'' (so the new task is available to all users on the system).
For developers there is a simple mechanism for version control and options to run external scripts to perform checks on the system before installing the task. It is also possible to access the ``install'' and ``uninstaller'' functions from the command line, via the ccp4ish -install and ccp4ish -uninstall options, which allows it to be incorporated into Makefiles or installation scripts for other packages.
The task installer can be accessed from the main CCP4i window via the ``System Administration->Install Tasks'' option.
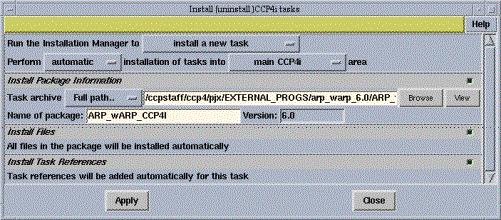
Figure 3: screenshot of the Task Installer interface
Two trial initiatives for automating tasks within CCP4i are included in 1.3.8, both of which involve passing information via XML files.
Background: before running the Molrep task it is useful to know certain details such as the number of monomers expected in the unit cell, and the existance of pseudotranslation vectors. These can be determined using the ``Cell Content Analysis'' and ``Analyse MR Data'' tasks respectively from the Molecular Replacement module, but in each case the data must be manually transfered from the log files to the Molrep interface.
In this pilot project, both the ``Cell Content Analysis'' and ``Analyse MR Data'' tasks generate XML output files recording key information. These files are generated by calls to the CCP4 XML-writing library PXXML within the MATTHEWS_COEFF and PEAKMAX programs, activated using the XMLOUT keyword in each.
The Molrep task can then automatically check for the existence of these output files and use the information in them to fill out the appropriate fields in the task interface for the user, reducing manual handling and typographical errors.
Reading of the XML files is performed using a set of utility functions built upon xml.tcl 1.9 and sgml.tcl 1.7, both included in CCP4i 1.3.8. (Note that the XML output of ``Cell Content Analysis'' has to be processed, with the number of monomers used being estimated as that number giving a solvent fraction closest to 50% of the unit cell.)
This option is turned off by default in CCP4i 1.3.8. Users wishing to try it can switch on the functionality in the ``XML Output'' folder in their ``Preferences'' (accessed from the menu on the RHS of the main window).
Background: occasionally when merging together MTZ files using CAD, it is possible that some will have different indexing conventions to the others leading to errors when combining them.
The CAD task now includes the option to ``Automatically check and enforce consistent indexing between files''. With this option selected each MTZ file is checked for consistent indexing against the first file, which is used as a reference. Files which are differently indexed are then reindexed prior to being merged. This mechanism cuts down on the overhead of manually diagnosing and correcting such cases when they arise.
This option uses XML passing within a single task rather than between tasks, as in the previous example. In this case ALMN is used to diagnose whether reindexing is required between two files, and if so then which reindexing operator to use. This information is written to a temporary XML file using the same mechanism as before, and is read from within the script using Tcl XML utilties.
In both these examples it would also have been possible to pass the same information using other mechanisms, for example by processing the log files directly using a variant on ``grep'' and other Unix-type cutting-and-sorting tools. However such methods are usually overly-complicated and prone to being easily broken by even small changes to log file formats. In contrast XML parameter passing is far simpler, more robust and easily extensible.
As of CCP4i 1.3.8 the interface source code includes inline ``doc-comments'', which are extracted and turned into html documentation of the code. Both the commented code and the extracted documentation are included in the current release, and will be useful for any programmers wishing to make CCP4i work more easily with their programs.
A number of longer-term projects are also envisaged:
The MTZ files impose a formal hierarchical structure on the reflection data they store, of the form ``Crystal->Dataset->Column'' (see Martyn Winn's article ``Development of the CCP4 software library'' in the newsletter 40, March 2002, for more explanation). In the future it should be possible to write programs which exploit this hierarchy for automate selection of data columns based only on a crystal or dataset name.
The MTZ viewer will display the crystal/dataset/column structure as a hierarchy or ``tree'', making it easier to visualise. Initially it is intended that the viewer should also act as a selection tool for datasets and columns, although ultimately it could also be used as an interface to perform CAD-like operations, for example to merge or split datasets.
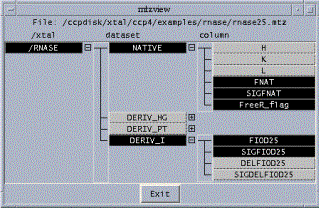
Figure 4: screenshot of prototype MTZ Hierarchical Viewer
The Project History Database is one of the most useful features of CCP4i above the ability to run the programs, as it tracks the jobs and associated datafiles which have been run within each project and allows the data from various parts of the structure determination to be accessed quickly. (For an overview see the article on ``Using CCP4i as a Project Management Tool'' in newsletter 38, April 2000.)
There are however a number of limitations in the current implementation, which spring from the fact that the code for handling the information in the project history database is embedded within the main CCP4i process. It is intended therefore to separate this component - the ``project history database handler'' or db handler for short - into an independent server process which can talk to the main CCP4i process via sockets.
In the short term this should not affect users at all; it will make a number of possibilities more feasible in the future. External packages such as MOSFLM would be able to interact with the database independently of CCP4i and leave records of jobs that it ran. The db handler could one day use a different database backend, for example a mySQL database, and interact with other databases storing different information, for example laboratory information management systems (LIMS).
Socket communications are also good for transfering information across networks, and so the db handler could run on a different computer to that running CCP4i. This would facilitate the transfer of CCP4i to a distributed computing environment, such as that envisioned in The Grid (for more information about Grid technologies see for example the Global Grid Forum website at http://www.gridforum.org/).
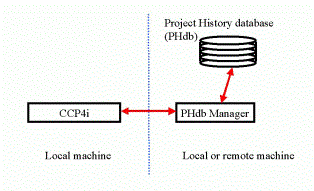
Figure 5: schematic for database interactions usin the db handler
Recent developments suggest that the Python scripting language is rapidly gaining ground as the language of choice for writing crystallographic computing applications. It is already being used in a number of successful high-profile projects, and future CCP4 programs are likely to involve a significant Python scripting component. (For more information about Python see e.g. http://www.python.org/).
At present CCP4i can run scripts directly only if they are written in Tcl. Extending the interface to allow run scripts written in Python enables CCP4i to keep up with these new developments as the new Python-based applications begin to emerge.
Currently, harvesting files are created in mmCIF format by a number of CCP4 programs, and store certain information about the dataset being processed which can be used later in the deposition process (see the CCP4 Harvesting documentation for more information).
Currently there is no way of tracking and validating these files during the structure solution process. A Harvesting File Manager is now under development which will allow users to track and validate the harvesting files, with the ultimate purpose being to make the deposition process simpler and faster.
The SAPHIRE task interface was designed to bring protein sequence information into CCP4 at an earlier stage of structure solution. Currently, protein sequences can be downloaded via this interface in FASTA format and edited if necessary before being saved locally. Extensions are being carried out to redesign the layout of this task and also to provide a graphical interface to running a local copy of CLUSTALW. (Note that CLUSTALW is not distributed by CCP4.)
CCP4i was originally developed by Liz Potterton, and Liz contributed the in-line documentation and the python run script functionality.
Pryank Patel is developing the GET_PROT/SAPHIRE application and the Data Harvesting Management tool. The BEAST interface was developed by Anne Baker with contributions from Peter Briggs. The ACORN and WHAT_CHECK interfaces were developed by Maria Turkenburg with the assistance of others including Yao Jia-xing, Eleanor Dodson, Gert Vriend, Liz Potterton and Peter Briggs. Other new interfaces were developed by Peter Briggs and Martyn Winn.
The automation projects were implemented by Martyn Winn, Alun Ashton and Peter Briggs.
Peter Briggs is responsible for the MapSlicer, Task Installer, MTZ Viewer and Project History Database developments. CCP4i is now maintained and developed by the DL CCP4 staff and other fixes and developments are due to them. Please send questions, requests and bug reports to us at ccp4@ccp4.ac.uk.