
 ) weighted by Fobs**2, which corresponds to the closeness of the maps calculated with Fobs and with exact or calculated phases.
) weighted by Fobs**2, which corresponds to the closeness of the maps calculated with Fobs and with exact or calculated phases.
The current techniques for ab initio macromolecular phasing are applicable at very low resolution and need to be followed by some phase extension procedures in order to improve the obtained image. Density modification methods can be used for this image improvement. Since they are usually applied at medium and high resolution (d < approx. 6 Å), a preliminary analysis of the applicability of these methods at very low resolution becomes necessary and is reported here.
While direct methods for small molecules have become an usual crystallographic tool, the solution of the phase problem for large macromolecules needs extra information like heavy atom derivatives or anomalous dispersion. During recent years some progress in the development of ab initio phasing methods at very low resolution has been reported (see, for example, Lunin, Urzhumtsev and Skovoroda, 1990; Subbiah, 1993; Harris, 1995; Volkman et al., 1995; Lunin et al., 1995, Urzhumtsev & Podjarny, 1995a, Andersson & Hovmöller, 1996; see also the review by Podjarny & Urzhumtsev, 1997). In particular, these methods have obtained the first crystallographic image for the 50S ribosome particle from Thermus thermophilus (Volkman et al., 1995; Urzhumtsev, Vernoslova and Podjarny, 1996). However, the resolution which can be obtained today by these methods is not high enough. A possible way to get a higher resolution image is to develop further the methods like FAM (Lunin et al., 1998; its development and application to the T50S ribosome data allowed an increase in the resolution from 100 Å initially to about 30 Å). Another way is to create or to modify existing methods for image improvement. These methods have been developed to be applied at "usual" resolution (2-5 Å) therefore at very low resolution some basic hypotheses may be not fulfilled. A special analysis should be done in order to show how these methods should be updated, if possible, to be applied at very low resolution data.
One of the basic groups of methods for image improvement is density modification (see, for example, Podjarny, Rees and Urzhumtsev, 1997). The usual iteration procedure of modification of the density distribution function, depending either on the density value in a given point or on the coordinates of this point, can be eventually applied at any resolution. The simplest information which can be used is the flat density in the solvent region (Bricogne, 1974). The following basic questions should be analysed :
Test calculations have been carried out using model data calculated from a density modulated envelope for the 50S ribosomal particle (Berkovich-Yellin, Wittmann & Yonath, 1990) arbitrarily placed in the experimentally observed unit cell (space group P43212; a = b = 496Å, c = 196Å) as described before in Lunin et al. (1995). Since no molecular model was used in the procedure, the contribution of the disordered solvent (Urzhumtsev & Podjarny, 1995b) was not taken into consideration. No experimental error was introduced.
The following procedure has been applied :
We considered that "good" structure factors at the starting resolution indicate the applicability of such flat solvent hypothesis (or flat envelope hypothesis), and that "good" phases at higher resolution indicate the possibility to use such procedure for image improvement. The limit of "goodness of added phases" was taken as 60 degrees (mean cosine value is 0.5) following Lunin & Woolfson (1993). Note that no iterations were done, all numbers are shown for structure factors obtained after a single density modification.
The procedure was applied for the maps of the resolution of 60, 40, 30 Å. Tables 1-3 give the results of the comparison of structure factors. Different columns correspond to different cut-off level values varied from 10 to 90% (percentage of the volume above the threshold value).
The amplitude correlation was calculated for Fobs (simulated data set) and Fcalc (after density modification) around their mean values.
The weighted phase correlation was calculated as the mean value of cos(
 ) weighted by Fobs**2, which corresponds to the closeness of the maps calculated with Fobs and with exact or calculated phases.
) weighted by Fobs**2, which corresponds to the closeness of the maps calculated with Fobs and with exact or calculated phases.
Cells are coloured accordingly to the criteria value; for the phase difference the limit value is 60 degrees. The shell of reciprocal space corresponding to the resolution slightly higher than the starting one is analysed in more detail and is given at the bottom of every table.
|
N |
Dmin |
Dmax |
refl |
10 |
20 |
30 |
40 |
50 |
60 |
70 |
80 |
90 |
|
1 |
80.0 |
500. |
40 |
91 |
96 |
92 |
87 |
83 |
|
|
|
|
|
2 |
60. |
80. |
49 |
83 |
88 |
89 |
84 |
74 |
|
|
|
|
|
3 |
50. |
60. |
52 |
29 |
16 |
-2 |
-2 |
5 |
|
|
|
|
|
4 |
45. |
50. |
48 |
1 |
-21 |
11 |
20 |
-3 |
|
|
|
|
|
5 |
40. |
45. |
74 |
7 |
-6 |
-15 |
0 |
4 |
|
|
|
|
|
6 |
35. |
40. |
118 |
- 4 |
11 |
-5 |
-13 |
-18 |
|
|
|
|
|
7 |
30. |
35. |
211 |
- 3 |
-13 |
-12 |
-7 |
3 |
|
|
|
|
|
8 |
25. |
30. |
383 |
10 |
5 |
6 |
3 |
7 |
|
|
|
|
|
9 |
20. |
25. |
876 |
10 |
14 |
6 |
9 |
7 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3.1 |
55. |
60. |
22 |
36 |
5 |
-1 |
7 |
1 |
|
|
|
|
|
3.2 |
50. |
55. |
30 |
-7 |
0 |
-5 |
-5 |
12 |
|
|
|
|
|
N |
Dmin |
Dmax |
refl |
10 |
20 |
30 |
40 |
50 |
60 |
70 |
80 |
90 |
|
1 |
80.0 |
500. |
40 |
98 |
99 |
97 |
92 |
90 |
|
|
|
|
|
2 |
60. |
80. |
49 |
96 |
97 |
96 |
93 |
85 |
|
|
|
|
|
2 |
50. |
60. |
52 |
38 |
55 |
46 |
17 |
-33 |
|
|
|
|
|
2 |
45. |
50. |
48 |
29 |
3 |
-13 |
-42 |
-44 |
|
|
|
|
|
2 |
40. |
45. |
74 |
51 |
40 |
11 |
-17 |
-46 |
|
|
|
|
|
2 |
35. |
40. |
118 |
2 |
23 |
15 |
-7 |
-13 |
|
|
|
|
|
2 |
30. |
35. |
211 |
27 |
21 |
1 |
-18 |
-30 |
|
|
|
|
|
2 |
25. |
30. |
383 |
21 |
11 |
-7 |
-26 |
-23 |
|
|
|
|
|
2 |
20. |
25. |
876 |
-5 |
0 |
-10 |
-3 |
12 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
2 |
55. |
60. |
22 |
40 |
56 |
55 |
20 |
-55 |
|
|
|
|
|
2 |
50. |
55. |
30 |
16 |
52 |
39 |
16 |
-16 |
|
|
|
|

 , in degrees
, in degrees|
N |
Dmin |
Dmax |
refl |
10 |
20 |
30 |
40 |
50 |
60 |
70 |
80 |
90 |
|
1 |
80.0 |
500. |
40 |
27 |
26 |
20 |
36 |
42 |
|
|
|
|
|
2 |
60. |
80. |
49 |
27 |
23 |
31 |
35 |
41 |
|
|
|
|
|
3 |
50. |
60. |
52 |
75 |
60 |
65 |
85 |
105 |
|
|
|
|
|
4 |
45. |
50. |
48 |
78 |
79 |
88 |
86 |
95 |
|
|
|
|
|
5 |
40. |
45. |
74 |
66 |
65 |
76 |
95 |
115 |
|
|
|
|
|
6 |
35. |
40. |
118 |
83 |
83 |
83 |
96 |
98 |
|
|
|
|
|
7 |
30. |
35. |
211 |
78 |
82 |
94 |
101 |
101 |
|
|
|
|
|
8 |
25. |
30. |
383 |
81 |
85 |
94 |
100 |
99 |
|
|
|
|
|
9 |
20. |
25. |
876 |
91 |
92 |
96 |
90 |
85 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3.1 |
55. |
60. |
22 |
62 |
43 |
49 |
81 |
121 |
|
|
|
|
|
3.2 |
50. |
55. |
30 |
85 |
72 |
77 |
87 |
93 |
|
|
|
|
|
N |
Dmin |
Dmax |
refl |
10 |
20 |
30 |
40 |
50 |
60 |
70 |
80 |
90 |
|
1 |
80.0 |
500. |
40 |
68 |
87 |
94 |
98 |
99 |
100 |
100 |
100 |
100 |
|
2 |
60. |
80. |
49 |
76 |
86 |
91 |
94 |
96 |
98 |
99 |
100 |
100 |
|
3 |
50. |
60. |
52 |
-7 |
4 |
14 |
28 |
33 |
33 |
33 |
31 |
25 |
|
4 |
45. |
50. |
48 |
18 |
20 |
15 |
11 |
12 |
15 |
19 |
22 |
22 |
|
5 |
40. |
45. |
74 |
-7 |
-3 |
5 |
4 |
-5 |
-5 |
0 |
1 |
4 |
|
6 |
35. |
40. |
118 |
4 |
6 |
3 |
2 |
3 |
1 |
0 |
-2 |
-2 |
|
7 |
30. |
35. |
211 |
-11 |
25 |
8 |
27 |
24 |
10 |
27 |
6 |
7 |
|
8 |
25. |
30. |
383 |
6 |
23 |
17 |
13 |
1 |
3 |
14 |
18 |
12 |
|
9 |
20. |
25. |
876 |
20 |
9 |
16 |
16 |
14 |
14 |
18 |
17 |
12 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3.1 |
55. |
60. |
22 |
|
0 |
3 |
10 |
12 |
12 |
12 |
12 |
10 |
|
3.2 |
50. |
55. |
30 |
|
7 |
17 |
32 |
38 |
36 |
34 |
34 |
28 |
|
N |
Dmin |
Dmax |
refl |
10 |
20 |
30 |
40 |
50 |
60 |
70 |
80 |
90 |
|
1 |
80.0 |
500. |
40 |
92 |
97 |
99 |
100 |
100 |
100 |
100 |
100 |
100 |
|
2 |
60. |
80. |
49 |
88 |
95 |
98 |
99 |
99 |
100 |
100 |
100 |
100 |
|
3 |
50. |
60. |
52 |
7 |
24 |
42 |
54 |
59 |
60 |
61 |
61 |
54 |
|
4 |
45. |
50. |
48 |
37 |
47 |
52 |
50 |
48 |
47 |
47 |
44 |
40 |
|
5 |
40. |
45. |
74 |
-10 |
20 |
35 |
40 |
33 |
25 |
21 |
20 |
19 |
|
6 |
35. |
40. |
118 |
-3 |
13 |
30 |
32 |
28 |
27 |
25 |
19 |
11 |
|
7 |
30. |
35. |
211 |
1 |
25 |
42 |
24 |
15 |
21 |
4 |
5 |
1 |
|
8 |
25. |
30. |
383 |
-5 |
27 |
34 |
18 |
1 |
-7 |
-9 |
-9 |
-5 |
|
9 |
20. |
25. |
876 |
11 |
1 |
-4 |
-15 |
-15 |
-10 |
-4 |
-3 |
2 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3.1 |
55. |
60. |
22 |
|
49 |
64 |
69 |
70 |
71 |
71 |
71 |
70 |
|
3.2 |
50. |
55. |
30 |
|
4 |
17 |
29 |
35 |
36 |
37 |
37 |
35 |

 , in degrees
, in degrees|
N |
Dmin |
Dmax |
refl |
10 |
20 |
30 |
40 |
50 |
60 |
70 |
80 |
90 |
|
1 |
80.0 |
500. |
40 |
27 |
26 |
21 |
20 |
12 |
2 |
1 |
1 |
1 |
|
2 |
60. |
80. |
49 |
32 |
25 |
22 |
18 |
16 |
12 |
10 |
3 |
1 |
|
3 |
50. |
60. |
52 |
87 |
79 |
67 |
62 |
55 |
53 |
51 |
59 |
72 |
|
4 |
45. |
50. |
48 |
80 |
69 |
69 |
69 |
64 |
66 |
69 |
67 |
67 |
|
5 |
40. |
45. |
74 |
96 |
79 |
63 |
59 |
68 |
70 |
73 |
79 |
84 |
|
6 |
35. |
40. |
118 |
88 |
89 |
82 |
77 |
78 |
79 |
83 |
83 |
85 |
|
7 |
30. |
35. |
211 |
90 |
83 |
71 |
91 |
90 |
87 |
90 |
92 |
93 |
|
8 |
25. |
30. |
383 |
92 |
78 |
80 |
83 |
88 |
94 |
94 |
94 |
96 |
|
9 |
20. |
25. |
876 |
86 |
88 |
96 |
97 |
93 |
91 |
89 |
88 |
89 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3.1 |
55. |
60. |
22 |
|
58 |
42 |
38 |
36 |
35 |
34 |
41 |
48 |
|
3.2 |
50. |
55. |
30 |
|
94 |
85 |
80 |
69 |
66 |
64 |
70 |
80 |
|
N |
Dmin |
Dmax |
refl |
10 |
20 |
30 |
40 |
50 |
60 |
70 |
80 |
90 |
|
1 |
80.0 |
500. |
40 |
91 |
98 |
92 |
89 |
|
|
|
|
|
|
2 |
60. |
80. |
49 |
93 |
89 |
87 |
81 |
|
|
|
|
|
|
3 |
50. |
60. |
52 |
88 |
92 |
79 |
71 |
|
|
|
|
|
|
4 |
45. |
50. |
48 |
63 |
65 |
53 |
55 |
|
|
|
|
|
|
5 |
40. |
45. |
74 |
59 |
72 |
60 |
64 |
|
|
|
|
|
|
6 |
35. |
40. |
118 |
16 |
14 |
5 |
19 |
|
|
|
|
|
|
7 |
30. |
35. |
211 |
10 |
3 |
6 |
17 |
|
|
|
|
|
|
8 |
25. |
30. |
383 |
24 |
2 |
9 |
28 |
|
|
|
|
|
|
9 |
20. |
25. |
876 |
8 |
15 |
15 |
9 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
6.1 |
37. |
40. |
66 |
9 |
2 |
15 |
15 |
|
|
|
|
|
|
6.2 |
35. |
37. |
52 |
24 |
25 |
-1 |
23 |
|
|
|
|
|
|
N |
Dmin |
Dmax |
refl |
10 |
20 |
30 |
40 |
50 |
60 |
70 |
80 |
90 |
|
1 |
80.0 |
500. |
40 |
99 |
99 |
96 |
94 |
|
|
|
|
|
|
2 |
60. |
80. |
49 |
98 |
98 |
96 |
95 |
|
|
|
|
|
|
3 |
50. |
60. |
52 |
95 |
96 |
93 |
89 |
|
|
|
|
|
|
4 |
45. |
50. |
48 |
91 |
93 |
90 |
85 |
|
|
|
|
|
|
5 |
40. |
45. |
74 |
86 |
96 |
90 |
85 |
|
|
|
|
|
|
6 |
35. |
40. |
118 |
54 |
44 |
2 |
-50 |
|
|
|
|
|
|
7 |
30. |
35. |
211 |
49 |
16 |
-22 |
-55 |
|
|
|
|
|
|
8 |
25. |
30. |
383 |
29 |
15 |
-22 |
-36 |
|
|
|
|
|
|
9 |
20. |
25. |
876 |
9 |
-18 |
-30 |
-21 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
6.1 |
37. |
40. |
66 |
48 |
53 |
8 |
-45 |
|
|
|
|
|
|
6.2 |
35. |
37. |
52 |
61 |
32 |
-8 |
-55 |
|
|
|
|
|

 , in degrees
, in degrees|
N |
Dmin |
Dmax |
refl |
10 |
20 |
30 |
40 |
50 |
60 |
70 |
80 |
90 |
|
1 |
80.0 |
500. |
40 |
23 |
15 |
21 |
43 |
|
|
|
|
|
|
2 |
60. |
80. |
49 |
27 |
22 |
28 |
29 |
|
|
|
|
|
|
3 |
50. |
60. |
52 |
29 |
21 |
22 |
34 |
|
|
|
|
|
|
4 |
45. |
50. |
48 |
30 |
24 |
35 |
42 |
|
|
|
|
|
|
5 |
40. |
45. |
74 |
38 |
26 |
37 |
38 |
|
|
|
|
|
|
6 |
35. |
40. |
118 |
65 |
74 |
88 |
115 |
|
|
|
|
|
|
7 |
30. |
35. |
211 |
65 |
85 |
100 |
114 |
|
|
|
|
|
|
8 |
25. |
30. |
383 |
79 |
84 |
99 |
107 |
|
|
|
|
|
|
9 |
20. |
25. |
876 |
87 |
94 |
106 |
99 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
6.1 |
37. |
40. |
66 |
63 |
72 |
81 |
113 |
|
|
|
|
|
|
6.2 |
35. |
37. |
52 |
67 |
75 |
96 |
117 |
|
|
|
|
|
|
N |
Dmin |
Dmax |
refl |
10 |
20 |
30 |
40 |
50 |
60 |
70 |
80 |
90 |
|
1 |
80.0 |
500. |
40 |
76 |
91 |
97 |
99 |
99 |
100 |
100 |
100 |
100 |
|
2 |
60. |
80. |
49 |
85 |
95 |
98 |
99 |
99 |
100 |
100 |
100 |
100 |
|
3 |
50. |
60. |
52 |
72 |
90 |
97 |
99 |
99 |
100 |
100 |
100 |
100 |
|
4 |
45. |
50. |
48 |
51 |
77 |
90 |
95 |
96 |
97 |
98 |
99 |
99 |
|
5 |
40. |
45. |
74 |
41 |
68 |
83 |
90 |
94 |
96 |
98 |
99 |
99 |
|
6 |
35. |
40. |
118 |
34 |
37 |
40 |
40 |
40 |
40 |
39 |
36 |
29 |
|
7 |
30. |
35. |
211 |
18 |
24 |
34 |
35 |
34 |
32 |
29 |
28 |
31 |
|
8 |
25. |
30. |
383 |
19 |
36 |
42 |
38 |
35 |
34 |
32 |
32 |
34 |
|
9 |
20. |
25. |
876 |
20 |
14 |
14 |
14 |
12 |
11 |
11 |
12 |
13 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
6.1 |
37. |
40. |
66 |
26 |
29 |
36 |
39 |
38 |
35 |
31 |
24 |
13 |
|
6.2 |
35. |
37. |
52 |
42 |
47 |
49 |
49 |
50 |
52 |
53 |
54 |
55 |
|
N |
Dmin |
Dmax |
refl |
10 |
20 |
30 |
40 |
50 |
60 |
70 |
80 |
90 |
|
1 |
80.0 |
500. |
40 |
95 |
99 |
100 |
100 |
100 |
100 |
100 |
100 |
100 |
|
2 |
60. |
80. |
49 |
95 |
98 |
100 |
100 |
100 |
100 |
100 |
100 |
100 |
|
3 |
50. |
60. |
52 |
79 |
94 |
99 |
100 |
100 |
100 |
100 |
100 |
100 |
|
4 |
45. |
50. |
48 |
78 |
94 |
98 |
99 |
100 |
100 |
100 |
100 |
100 |
|
5 |
40. |
45. |
74 |
69 |
89 |
96 |
98 |
99 |
99 |
100 |
100 |
100 |
|
6 |
35. |
40. |
118 |
57 |
69 |
77 |
80 |
81 |
79 |
76 |
72 |
66 |
|
7 |
30. |
35. |
211 |
51 |
66 |
70 |
70 |
70 |
68 |
66 |
62 |
56 |
|
8 |
25. |
30. |
383 |
31 |
50 |
52 |
46 |
43 |
42 |
40 |
36 |
31 |
|
9 |
20. |
25. |
876 |
30 |
28 |
1 |
-18 |
-23 |
-24 |
-25 |
-25 |
-24 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
6.1 |
37. |
40. |
66 |
53 |
68 |
77 |
81 |
81 |
79 |
74 |
69 |
61 |
|
6.2 |
35. |
37. |
52 |
61 |
70 |
77 |
80 |
80 |
79 |
78 |
76 |
72 |

 , in degrees
, in degrees|
N |
Dmin |
Dmax |
refl |
10 |
20 |
30 |
40 |
50 |
60 |
70 |
80 |
90 |
|
1 |
80.0 |
500. |
40 |
34 |
20 |
15 |
7 |
3 |
3 |
1 |
1 |
1 |
|
2 |
60. |
80. |
49 |
35 |
21 |
17 |
10 |
9 |
7 |
4 |
3 |
2 |
|
3 |
50. |
60. |
52 |
48 |
31 |
18 |
11 |
9 |
8 |
7 |
7 |
1 |
|
4 |
45. |
50. |
48 |
48 |
26 |
20 |
10 |
8 |
6 |
5 |
5 |
1 |
|
5 |
40. |
45. |
74 |
52 |
38 |
26 |
15 |
12 |
10 |
7 |
5 |
3 |
|
6 |
35. |
40. |
118 |
71 |
63 |
58 |
57 |
54 |
53 |
54 |
57 |
66 |
|
7 |
30. |
35. |
211 |
64 |
59 |
54 |
52 |
55 |
55 |
57 |
60 |
65 |
|
8 |
25. |
30. |
383 |
78 |
72 |
68 |
69 |
74 |
75 |
77 |
78 |
82 |
|
9 |
20. |
25. |
876 |
78 |
82 |
93 |
101 |
102 |
102 |
101 |
101 |
100 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
6.1 |
37. |
40. |
66 |
71 |
62 |
59 |
57 |
51 |
49 |
53 |
56 |
73 |
|
6.2 |
35. |
37. |
52 |
70 |
64 |
56 |
56 |
57 |
58 |
57 |
59 |
58 |
|
N |
Dmin |
Dmax |
refl |
5 |
10 |
20 |
30 |
40 |
50 |
60 |
70 |
80 |
90 |
|
1 |
80.0 |
500. |
40 |
73 |
85 |
96 |
99 |
100 |
100 |
100 |
100 |
100 |
100 |
|
2 |
60. |
80. |
49 |
71 |
87 |
97 |
99 |
99 |
100 |
100 |
100 |
100 |
100 |
|
3 |
50. |
60. |
52 |
49 |
76 |
92 |
97 |
98 |
99 |
99 |
100 |
100 |
100 |
|
4 |
45. |
50. |
48 |
50 |
69 |
87 |
95 |
96 |
97 |
98 |
99 |
99 |
100 |
|
5 |
40. |
45. |
74 |
51 |
68 |
89 |
96 |
97 |
98 |
99 |
99 |
99 |
100 |
|
6 |
35. |
40. |
118 |
67 |
77 |
91 |
96 |
98 |
98 |
99 |
99 |
100 |
100 |
|
7 |
30. |
35. |
211 |
65 |
76 |
87 |
92 |
95 |
97 |
98 |
99 |
99 |
100 |
|
8 |
25. |
30. |
383 |
41 |
48 |
56 |
58 |
57 |
57 |
55 |
50 |
40 |
22 |
|
9 |
20. |
25. |
876 |
25 |
34 |
36 |
31 |
28 |
26 |
25 |
24 |
22 |
21 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
8.1 |
27. |
30. |
199 |
45 |
52 |
59 |
63 |
62 |
61 |
58 |
52 |
38 |
19 |
|
8.2 |
25. |
27. |
184 |
31 |
39 |
49 |
48 |
47 |
47 |
48 |
47 |
42 |
36 |
|
9.1 |
22. |
25. |
427 |
21 |
31 |
35 |
29 |
25 |
25 |
25 |
26 |
25 |
24 |
|
9.2 |
21. |
22. |
213 |
14 |
22 |
18 |
14 |
12 |
9 |
6 |
4 |
0 |
-2 |
|
N |
Dmin |
Dmax |
refl |
5 |
10 |
20 |
30 |
40 |
50 |
60 |
70 |
80 |
90 |
|
1 |
80.0 |
500. |
40 |
93 |
97 |
100 |
100 |
100 |
100 |
100 |
100 |
100 |
100 |
|
2 |
60. |
80. |
49 |
87 |
95 |
99 |
100 |
100 |
100 |
100 |
100 |
100 |
100 |
|
3 |
50. |
60. |
52 |
78 |
90 |
98 |
100 |
100 |
100 |
100 |
100 |
100 |
100 |
|
4 |
45. |
50. |
48 |
72 |
88 |
97 |
99 |
100 |
100 |
100 |
100 |
100 |
100 |
|
5 |
40. |
45. |
74 |
84 |
89 |
96 |
99 |
99 |
100 |
100 |
100 |
100 |
100 |
|
6 |
35. |
40. |
118 |
85 |
91 |
97 |
99 |
99 |
100 |
100 |
100 |
100 |
100 |
|
7 |
30. |
35. |
211 |
87 |
93 |
97 |
98 |
99 |
100 |
100 |
100 |
100 |
100 |
|
8 |
25. |
30. |
383 |
67 |
73 |
81 |
85 |
85 |
84 |
84 |
83 |
81 |
82 |
|
9 |
20. |
25. |
876 |
53 |
64 |
67 |
60 |
54 |
50 |
47 |
43 |
40 |
36 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
8.1 |
27. |
30. |
199 |
71 |
77 |
83 |
87 |
87 |
87 |
86 |
85 |
84 |
86 |
|
8.2 |
25. |
27. |
184 |
58 |
64 |
76 |
79 |
79 |
78 |
77 |
76 |
73 |
66 |
|
9.1 |
22. |
25. |
427 |
56 |
67 |
74 |
72 |
68 |
67 |
66 |
64 |
62 |
58 |
|
9.2 |
21. |
22. |
213 |
48 |
58 |
53 |
35 |
27 |
21 |
15 |
10 |
6 |
4 |

 , in degrees
, in degrees|
N |
Dmin |
Dmax |
refl |
5 |
10 |
20 |
30 |
40 |
50 |
60 |
70 |
80 |
90 |
|
1 |
80.0 |
500. |
40 |
35 |
25 |
20 |
7 |
1 |
1 |
1 |
1 |
1 |
0 |
|
2 |
60. |
80. |
49 |
44 |
35 |
25 |
7 |
5 |
2 |
2 |
2 |
1 |
1 |
|
3 |
50. |
60. |
52 |
48 |
40 |
24 |
10 |
8 |
7 |
7 |
4 |
4 |
4 |
|
4 |
45. |
50. |
48 |
51 |
39 |
24 |
13 |
10 |
9 |
8 |
8 |
5 |
4 |
|
5 |
40. |
45. |
74 |
43 |
33 |
24 |
15 |
10 |
8 |
7 |
6 |
4 |
2 |
|
6 |
35. |
40. |
118 |
50 |
36 |
21 |
11 |
9 |
8 |
7 |
5 |
1 |
2 |
|
7 |
30. |
35. |
211 |
35 |
38 |
21 |
17 |
14 |
10 |
9 |
6 |
4 |
3 |
|
8 |
25. |
30. |
383 |
57 |
53 |
44 |
43 |
43 |
43 |
44 |
46 |
49 |
54 |
|
9 |
20. |
25. |
876 |
66 |
60 |
60 |
66 |
69 |
71 |
72 |
73 |
73 |
75 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
8.1 |
27. |
30. |
199 |
53 |
52 |
42 |
42 |
42 |
42 |
43 |
45 |
48 |
54 |
|
8.2 |
25. |
27. |
184 |
62 |
55 |
46 |
43 |
43 |
44 |
46 |
47 |
50 |
53 |
|
9.1 |
22. |
25. |
427 |
63 |
55 |
50 |
53 |
55 |
58 |
59 |
60 |
60 |
64 |
|
9.2 |
21. |
22. |
213 |
67 |
63 |
65 |
76 |
81 |
82 |
83 |
85 |
86 |
88 |
The following observations can be made :
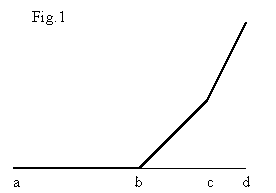 As was discussed by Lunin (1988), the basic idea which is behind most of density modification procedures and which defines the form of the density modification function is the closeness of the electron density histograms. In our case, the corresponding histograms have been calculated at different resolution from 20 to 90 Å. We have no possibility to discuss here the details of this analysis and can only mention that the density modification function corresponding to the optimal density modification from 90 to 20 Å resolution maps can be nicely approximated by a function schematically presented in Figure 1 : at the interval (a,b) it corresponds to the solvent flattening, at the interval (b,c) it "keeps" the density values and the interval (c,d) the function needs to sharpen the highest density values. In general, this function supports the idea of "soft" modification. The application of histogram-fitted density modification will be discussed elsewhere.
As was discussed by Lunin (1988), the basic idea which is behind most of density modification procedures and which defines the form of the density modification function is the closeness of the electron density histograms. In our case, the corresponding histograms have been calculated at different resolution from 20 to 90 Å. We have no possibility to discuss here the details of this analysis and can only mention that the density modification function corresponding to the optimal density modification from 90 to 20 Å resolution maps can be nicely approximated by a function schematically presented in Figure 1 : at the interval (a,b) it corresponds to the solvent flattening, at the interval (b,c) it "keeps" the density values and the interval (c,d) the function needs to sharpen the highest density values. In general, this function supports the idea of "soft" modification. The application of histogram-fitted density modification will be discussed elsewhere.