
This task allows you to run PHASER for molecular replacement, up to and including building multi-subunit complexes, each component of which is constructed from an ensemble of structures. Optionally, the model can be:
• Put through a few cycles of REFMAC refinement (option selected by default)
• Symmetry matched to a reference PDB
• Processed by running a coot script (e.g. to fill partial residues in a model that has been chainsawed).
Reflections
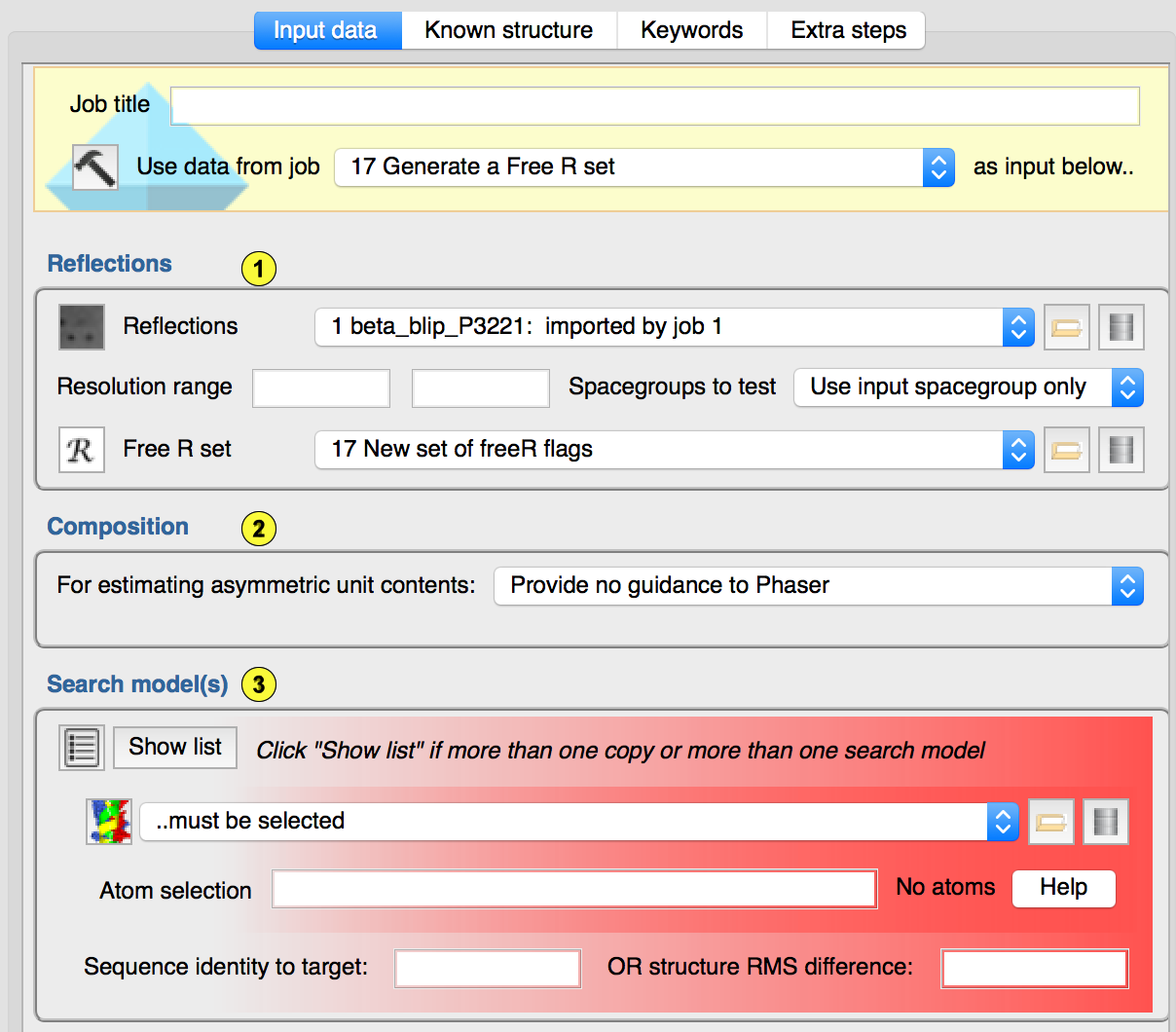
Reflections.(1.1) Hopefully self-explanatory.
Resolution range. Based on the expected LLGs, PHASER will try to search with the best resolution. You may override the maximum resolution that PHASER will attempt here.
Spacegroups. Where there is ambiguity, you can invite PHASER to attempt MR in more than one spacegroup: data reduction generally provides insight into the likely (but not certain!) pointgroup of the crystal, with less reliable guidance as to the space group.
Free R set. Although PHASER does not take advantage of a FreeR set, the optional REFMAC cycles which run at the end of this task should be given the FreeR data object that you are using for this crystal form.
Composition(1.2)
The maximum likelihood approach in PHASER works best if it knows the total scattering in the unit cell. The composition section allows you to provide that information in one of three ways. Firstly, you may choose to offer no information. In this case, PHASER will identify the most probable solvent content for the resolution of data provided, and infer the amount of scattering from that. Secondly, you may provide an estimate of the total molecuar weight of protein and/or nucleic acid in the crystal. In the event of non crystallographic symmetry, this will be more than the sum of the molecular weights of the macromolecular chains that you think you have crystallized. Phaser will evaluate the number you provide here and give feedback on the number of copies thereof that are most consistent with the probable solvent content. Thirdly, you can provide the sequences of all of the species you think to be present in the crystal together with the number of copies of each that you expect to form the crystallographic asymmetric unit. Again, PHASER will provide feedback on the number of copies of your estimated composition that are most likely based on the probable solvent content.Search model(s)(1.3)
Simple search with a single structure: If you have a single structure that you want to position in the unit cell, then all you have to do is provide that structure here :-). The user can specify a subset of the structure to search with using the "coordinate selection" syntax described here. This may be useful if the structure you are solving corresponds to one chain of a PDB that you have downloaded, or if there are segments of the structure that you think will be in a different conformation in the new crystal (typically useful for N- and C-terminii or for known flexible loops). The user must also specify either the sequence identity of the search model to the species present in the crystal, or the expected RMS coordinate similarity between them. These parameters are required for PHASER's maximum likelihood approach, although even reputable crystallographers occasionally just provide the number 0.9 as sequence identityMore complex MR search scenarios: Click the "Show list" button if you want to:
• Position multiple copies of a search molecule in the unit cell,
• Build up a macromolecular hetero-oligomeric complex, and/or
• Exploit ensembles (multiple superimposed search models for a particular component of the asymmetric unit)
This will cause the expanded search model dialog to be displayed, which provides control of the search models provided and the searches to be carried outExpanded Search Model Dialog
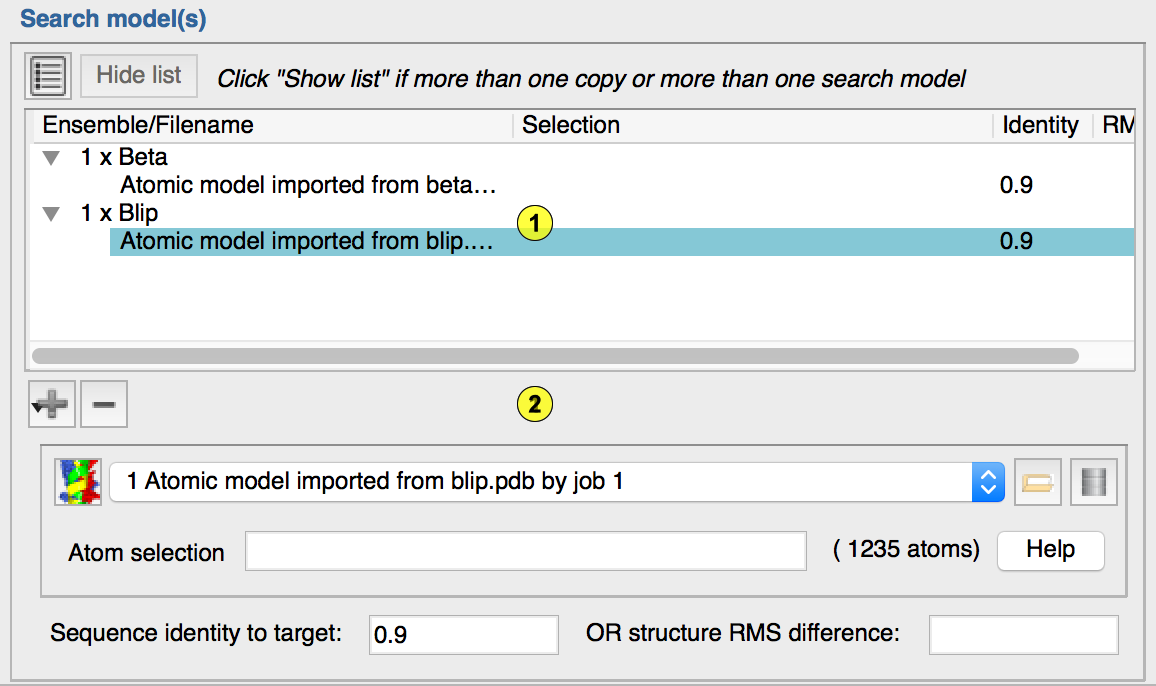
• The search model tree (2.1), and
• The search model detail (2.2)
The "search model tree" shows an overview of the search model(s) that you have provided to the gui. This is made up of "ensembles", each of which is made up of "structures". What is displayed in the "search model detail" depends on what is selected in the search model tree.Ensembles: An ensemble corresponds to a unit of structure that can be positioned in an MR run. It generally constitutes a "rigid body" that is likely to be common between the search model(s) provided and the structure for which the data have been collected. In the simplest case, an ensemble can comprise a single structure. If this works (which it will in > 90% of solvable cases), then more power to your elbow :-). When an ensemble is selected in the search model tree, the search model detail allows you to specify 1) The number of copies of the ensemble to search for in this run, and 2) A short one word alphanumeric identifier to associate with this ensemble.
Structures: If searching with a single structure in your ensemble fails, then it may be that adding more than one (superimposed) structure into your ensemble can help. The idea is that each superimposed structure will have different strengths and weaknesses, such that the search for the composite model (i.e. ensemble) may succeed where searching for each constituent structure fails. N.B. It is currently up to the user to superimpose the structures prior to giving them to CCP4i2. When a "structure" is selected in the search model tree, the search model detail allows/requires you to specify similar properties to those described above for a simple search model.
Adding and deleting ensembles and structures. To add an ensemble to the search, click "+" and select "Add ensemble". To add a structure into an ensemble, select the ensemble in the search model tree, click "+" and select "Add structure in ensemble". To delete an ensemble or structure, select it in the search model tree and click "-"
Searching for more than one copy. To search for more than one copy of an ensemble, you will need to have clisked "Show list" to reveal the Expanded search model dialog. The number of copies to search for in this run is set be selecting the relevant ensemble in the search model tree and then specifying the number to search fo r in the search model detail panel.