The final stage of the crystallographic structure solution is the deposition of the final structure and the observations in the Protein Data Bank (PDB). The PDB require that the files be provided in a standard form, specifically the observations and the atomic model must be in CIF format and and TLS contributions to the refinement calculation must be included. This task will automatically prepare the required files in the appropriate formats.
Input
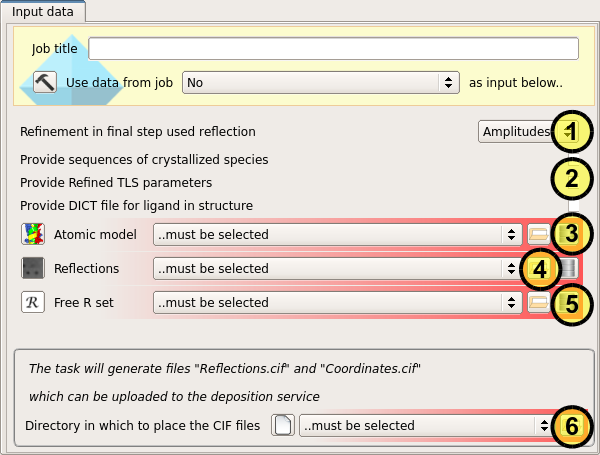
Firstly, the type of observations used for the final refinement must be specified, whether amplitudes or intensities (1).
Next, any optional files required to reproduce the final refinement must be specified. These may include the sequence data for the crystallized species, in case not all of the structure was built; the refined TLS parameters from the final refinement in the case of TLS refinement (most cases); and the dictionary for any ligands or other non-standard monomers in the crystal (2).
The refined atomic model (3), the observations against which the model was refined (4), and the Free-R set must then be specified so that the refinement result can be confirmed (5). The observations may be amplitudes or intensities as specified previously, but must be the same as used in the final refinement.
Finally a folder must be selected, into which the deposition files 'Reflections.cif' and 'Coordinates.cif' will be placed (6).
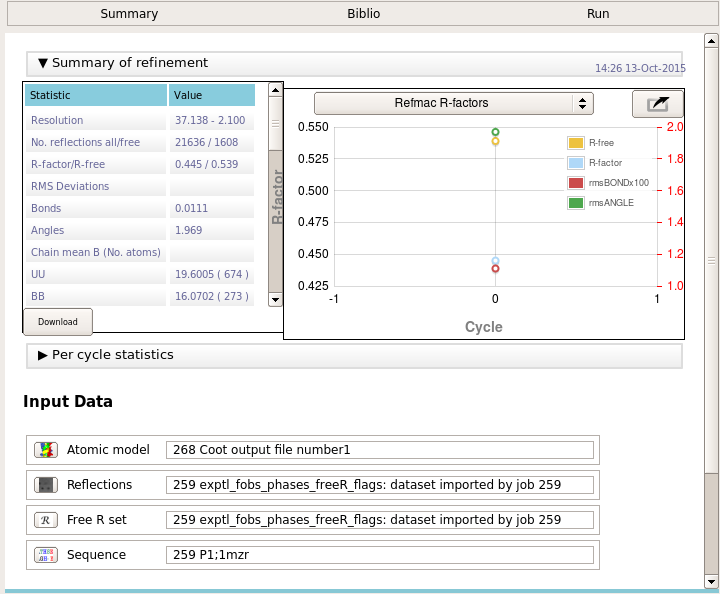
The deposition task will run refmac to try and reproduce the refinement statistics from the end of the structure refinement. You should check the refinement statistics to make sure that they match the final refinement cycle. If not, check that you have provided any necessary TLS and dictionary files.
The CIF files for deposition will be placed in the folder specified in the task input.