Metal Coordination Groups in Proteins
Some Comments on Geometry, Constitution and B-values
Marjorie Harding
Structural Biochemistry Group, Institute of Cell and Molecular Biology,
Michael Swann Building, University of Edinburgh, Edinburgh EH9 3JR
Metals are found in a wide variety of proteins where they may have important functional (usually catalytic) or structural roles. In a large proportion of structures the coordination group around the metal atom is made up of donor groups (carboxylate, imidazole, etc) from several amino-acid side chains, usually, but not necessarily, within one polypeptide chain; water molecules or other small molecules incorporated in the crystal as inhibitors, substrate analogues, cofactors etc. may also participate in the coordination group. In iron proteins, haem groups and clusters like Fe4S4 are also common.
The aim of recent work here (Harding, 1999, 2000, 2001) has been to provide information on metal coordination geometry which could be of use to protein crystallographers determining structures - at the stage of interpreting an electron density map, or in the restrained refinement of structures where the data is of limited resolution, or in the validation of structures. The work included a systematic extraction of geometric data from the Protein Data Bank (PDB, Bernstein et al., 1977; Berman et al., 2000) for metalloproteins of six selected metals. This note describes some further work on these metal coordination groups, which although not geometrical, might also be of relevance in protein structure determination - a) a systematic description and listing of metal coordination groups in terms of constituent amino-acid donor groups and their relative positions in the amino-acid sequence of the polypeptide chain, and b) a brief comparison of reported B values for the metal atom and the donor atoms in some of these groups. Much of this information has been assembled in a website on metal coordination groups. This includes links to some other websites relevant to metal coordination chemistry in proteins; among these is metalloscripps, which contains extensive geometrical information and tools for manipulation. The present descriptions and listings take no account of biological function or other properties of the metal sites; Degtyarenko (2000) describes an approach in terms of 'bioinorganic motifs' which does take function into account, and gives information on the available databases relevant to the field.
| 1ctt | resolution = 2.20; total no. atoms = 2286; no. metal atoms = 1 | ||||||||||||||||||
| metal no. |
coordn sphere |
donor | dist (A) |
dif from target |
occ. product |
B metal |
B donor | 1 ZN 296 | |||||||||||
| 1 |
| 2.02 | 0.02 | 1.0 | 22.4 | 26.3 | |||||||||||||
| 1 | 2.42 | 0.13 | 1.0 | 22.4 | 23.6 | ||||||||||||||
| 1 | 2.11 | -0.18 | 1.0 | 22.4 | 19.4 | ||||||||||||||
| 1 | 1.84 | -0.25 | 1.0 | 22.4 | 13.7 | ||||||||||||||
| cngroup is HCC with CN 4 Zn, sequence diffs 27 3 | |||||||||||||||||||
| COORDINATION NUMBER: 4 | |||||||||||||||||||
| nearest description of shape - tetrahedral (r.m.s. devn from tet 7.7, r.m.s. devn from sqp 41.1 degrees) | |||||||||||||||||||
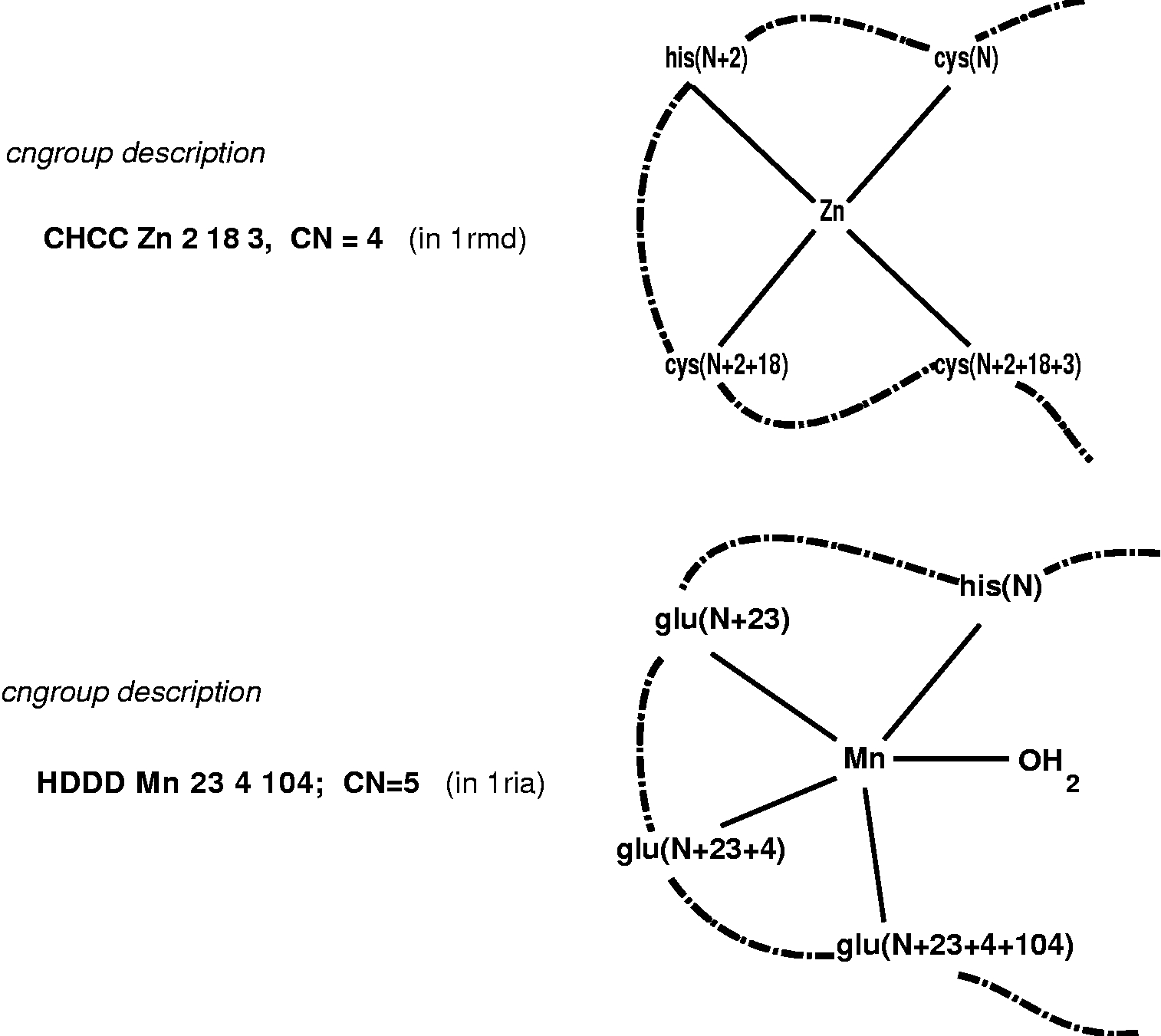
The description of a coordination group, or cngroup, used here includes the nature of the amino-acid donor groups, their sequence and separation (number of residues apart) in the protein chain(s), together with the metal coordination number (which includes non-protein donors, water molecules and other ligands). This information can be summarised as in the two examples below, where it is assumed that N is a residue number, cysteine coordinates through the thiolate sulphur, and histidine through imidazole nitrogen.

Methods, procedures: The basis for generating the cngroup information is the program MP (Harding 2001, Acta Cryst D57, ) which reads a PDB file, extracts coordinates and occupancy of each metal atom, and those of all atoms within 3.6 Å of the metal atom. Using target distances for each metal-donor atom combination it identifies atoms as donors if they lie within (target distance + tolerance) of the metal, and lists the amino-acids and residue numbers to which they belong, and the coordination number - as already illustrated in Table 1. It also evaluates sig, the r.m.s. deviation of the observed distances from the target values, and an alternative coordination number which would be found if an alternative, larger value of tolerance were used; these two, together with the resolution of the structure determination are useful in assessing the accuracy with which the cngroup has been identified. Metal coordination groups in which any atoms are disordered or have occupancy less than 0.7, are omitted.
Lists of PDB codes were obtained from the Jena Image Library search facility . (This gives a more complete listing than the PDB-3D Browser which searches in HET group names; some HET group names are odd, for example OC5 for Ca(OH2)52+, and in such cases no Ca atom is detected by the Browser. ) From these lists protein and protein-nucleic acid complexes were selected, with structures determined by diffraction to a resolution < 2.8 Å, and the program MP run, for all the structures available in the RCSB release of Feb2001. Additional smaller programs then gave the information on cngroup descriptions for the full lists or for selections from them. One such selection is a 'representative set' which excludes any structure which has more than 30% sequence identity with any other in the set; this used a culled PDB file 'cullpdb_pc30_res3.0_...'.
The list of cngroup descriptions is sorted in alphabetical order and gives sequence, metal, separation of coordinating residues in protein sequence, number of coordinating groups, pdbcode, 3 indicators of reliability, and the name of the metal and the first coordinated amino-acid in the chain. One letter amino-acid codes are used to specify the donor groups, and O indicates main chain carboxyl oxygen as donor. A carboxylate group (D or E) is always treated as one donor group, whether it is mono- or bidentate; at lower resolutions the distinction is not reliable, and particularly in Zn complexes, intermediate states are possible. The apaprent mono- or bi-dentate status is indicated at the end of the record. The presence of water molecules or donor atoms from non-protein molecules is indicated and an alternative output option includes these within the cngroup before sorting into sequence order .
Concerning cngroup definition: An atom is identified here as a donor when its distance from the metal atom is within (target distance + tolerance). The target distances have been carefully established using appropriate small molecule compounds in the CSD and checking against high resolution protein structures (Harding 1999, 2000, 2001). Errors in determination of atom positions, especially in low resolution structures might result in incorrect decisions on whether or not an atom is within the metal coordination group. For this reason structures determined at resolutions less good that 2.8 Å are not included at all. The tolerance was set at 0.75 Å after examining the distribution of (observed - target) distances. When the resolution is <1.8 Å there should be no 'wrong decisions' about whether an atom is within the metal coordination group; when the resolution is poorer, but still < 2.8 Å, some 'wrong decisions' will inevitably be made, but their number should be well under 5% of the whole. The three indicators given for each cngroup are provided to show more about the reliability of these decisions: i) the resolution, ii) the number of additional donor atoms which would have been found if the tolerance had been 0.95A, and iii) the r.m.s. deviation of observed from target distance in the cngroup.
A few metal atoms in the cngroup listings have coordination numbers lower than would normally be expected (i.e. <5 for Ca, <4 for Mg, Mn, Fe, Zn, <3 for Cu) . Usually this is the result of failure to identify a donor group such as a water molecule, in the electron density map, but in a few cases it could be the result of a shortcoming in the software, which does not (yet) detect when the metal atom is coordinated to a donor group in a neighbouring asymmetric unit of the crystal.
Lists generated and their possible uses: The lists now include all proteins containing any of the metals Ca, Mg, Mn, Cu, Zn, whose structures have been determined at resolution < 2.8 Å. They include many repeat entries where the same metalloprotein molecule occurs more than once in the crystal asymmetric unit, as well as occurrences of the same cngroup sequence in very closely related proteins such as mutants. They are sorted so that identical cngroups in different proteins appear next to each other. From these lists summary lists are also provided, which group together all protein structures which have the same cngroup sequence . Lists are also provided for a representative set of proteins (no two having sequence identity > 30%), and the summary lists for these - Table 2 is an example showing the form of the summary list.
| DDDN | Ca | 88 | 2 | 1 | . | . | . | . | 6 | 1 | 1alv |
| DDDOD | Ca | 2 | 2 | 2 | 5 | . | . | . | 6 | 1 | 2scp |
| DDDOE | Ca | 2 | 2 | 2 | 5 | . | . | . | 6 | 1 | 1cdl |
| DDDOE | Ca | 2 | 2 | 2 | 5 | . | . | . | 6 | 4 | 1acc 1sra 1vrk 2pvb |
| DDEOOD | Ca | 2 | 7 | 34 | 3 | 10 | . | . | 6 | 1 | 1acc |
| DDND | Ca | 2 | 5 | 1 | . | . | . | . | 6 | 1 | 2por |
| DDNNOE | Ca | 2 | 2 | . | 2 | 5 | . | . | 6 | 1 | 2cdl |
| DDNOOE | Ca | 2 | 2 | 2 | 2 | 3 | . | . | 6 | 1 | 1cdl |
| DDNOE | Ca | 2 | 2 | 2 | 5 | . | . | . | 5 | 1 | 1cdl |
| DDNOE | Ca | 2 | 2 | 2 | 5 | . | . | . | 6 | 2 | 1rec 1vrk |
| CCCC | Zn | 3 | 7 | 6 | . | . | . | . | 5 | 1 | 1zme |
| CCCC | Zn | 3 | 7 | 7 | . | . | . | . | 5 | 1 | 1hwt |
| CCCC | Zn | 3 | 14 | 3 | . | . | . | . | 4 | 1 | 1dsz |
| CCCC | Zn | 3 | 17 | 3 | . | . | . | . | 4 | 3 | 1dcq 1rmd 1zin |
| CCCC | Zn | 3 | 22 | 3 | . | . | . | . | 4 | 1 | 1zbd |
| HCC | Zn | 27 | 3 | . | . | . | . | . | 4 | 1 | 1ctt |
| HCCC | Zn | 11 | 1 | 10 | . | . | . | . | 4 | 1 | 1btk |
| HCCC | Zn | 13 | 10 | 3 | . | . | . | . | 4 | 1 | 1gpc |
| HCCC | Zn | 30 | 3 | 16 | . | . | . | . | 4 | 1 | 1ptq |
These lists may have a variety of uses. It would be quick and easy to check whether in a newly determined metalloprotein structure the constitution of the metal coordination group is the same as that in a known structure, or structures. Here we plan to make geometrical comparisons after selecting proteins which have the same sequence of coordinating amino-acids in the cngroup, or closely related sequences, even though the overall sequences of the proteins are very different; polypeptide chain conformations in the chelating loops and nearby regions of the structure will then be compared, either by examining sequences of torsion angles, or by graphical superposition. Several sets of cngroup descriptions are easily recognised as familiar motifs, e.g. Ca proteins with EF hands, some Zn fingers, etc., which suggests that these listings may have some use in classifying metalloprotein structures. A long term aim of this project is to establish a representative set (in constitution and geometry) of metal coordination groups in proteins; this would have considerable overlap with the list of metal coordination groups in a representative set of proteins, but not be identical to it. This aim bears some relationship, but is also complementary to work nearing completion at University College London (M.W.MacArthur - private communication). The project there, which is in a much more advanced stage than this one, involves building a library of structural motifs (of metal coordination sites) including 3 dimensional aspects. These motifs, which can be used as templates or probes for a systematic classification of sites, include the positions of donor atoms of constituent amino-acids relative to the metal, regardless of the positions in which the amino-acids occur in the polypeptide sequence.
Some statistics of the numbers of structures and sequences found are given in Table 3 . (A few of the results or sequences are trivial, e.g. the numerous examples of Mg(OH2)n2+ cations with no protein donor groups. There are also examples where groups are separately reported although the difference is small - e.g. a difference in coordination number due to different numbers of water molecules located near the metal.)
| all metalloproteins | Ca | Mg | Mn | Cu | Zn | |
| No. of structures | 1081 | 546 | 207 | 183 | 782 | |
| No. of cngroups including repeats within asymmetric unit | 2412 | 1113 | 571 | 434 | 1560 | |
| No. of cngroups occurring only once in different proteins | 381 | 146 | 102 | 67 | 301 | |
| No. of cngroups occurring in different proteins | 2 times | 69 | 41 | 24 | 16 | 54 |
| 3 times | 49 | 23 | 10 | 4 | 18 | |
| 4 times | 23 | 10 | 5 | 3 | 8 | |
| 5 times | 87 | 30 | 11 | 10 | 65 | |
| representative set of metalloproteins | Ca | Mg | Mn | Cu | Zn | |
| No. of structures | 121 | 127 | 21 | 22 | 116 | |
| No. of cngroups including repeats within asymmetric unit | 289 | 271 | 53 | 52 | 26 | |
| No. of cngroups occurring only once in different proteins | 184 | 78 | 29 | 31 | 183 | |
| No. of cngroups occurring in different proteins | 2 times | 10 | 7 | 0 | 0 | 8 |
| 3 times | 1 | 5 | 1 | 2 | 3 | |
| 4 times | 2 | 4 | 0 | 1 | 1 | |
| >5 times | 0 | 5 | 0 | 0 | 1 | |
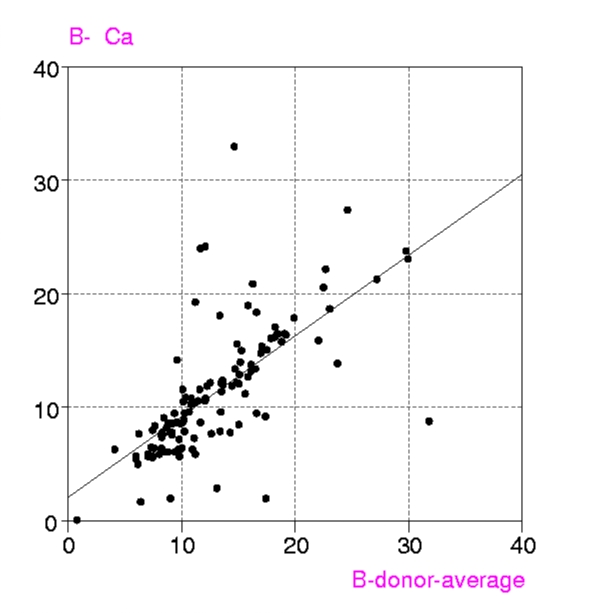
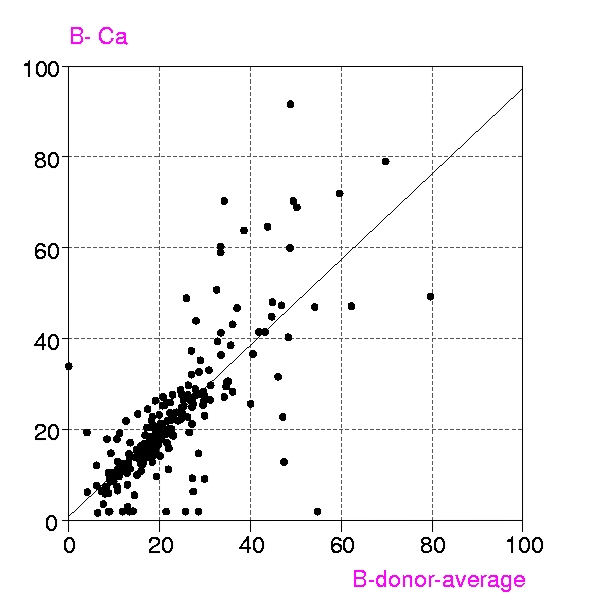
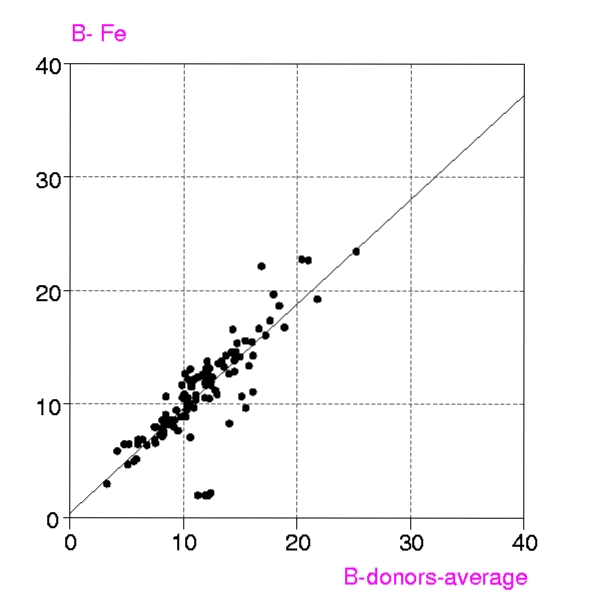
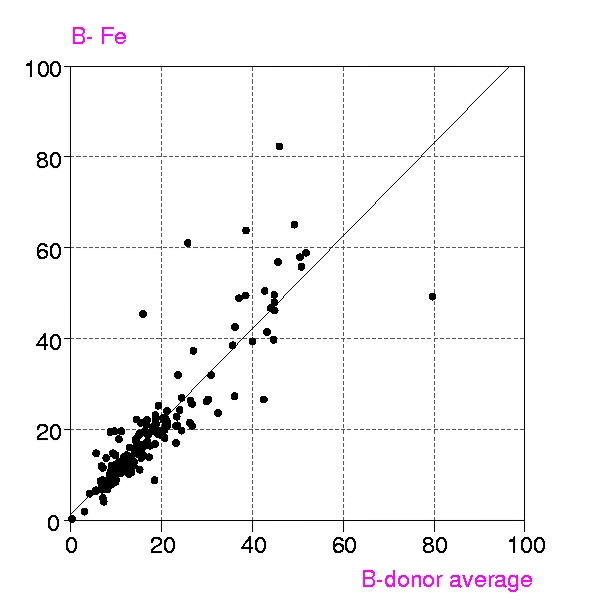
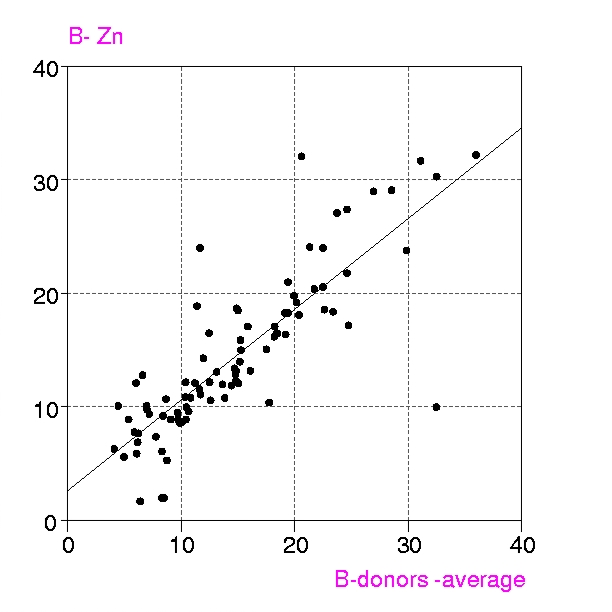
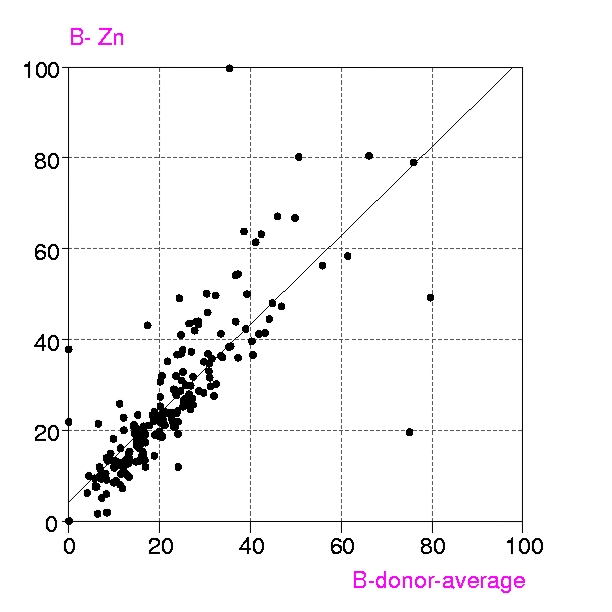
Procedures : Metalloproteins in the PDB up to July 1999 containing any of the metals Ca, Mg, Mn, Fe, Mn, Cu, Zn, have been examined in two groups, a) all structures with resolution < 1.6 Å and b) 'representative macromolecules' with resolution up to 2.8 Å. Coordination groups where the metal atom occupancy or any donor atom occupancy is less than 1.0 were excluded. The results were displayed in a spreadsheet (VISTA of the CSD system), and included, for each metal coordination group, Bmetal, the mean, minimum and maximum values of Bdonor, as well as the coordination number, resolution, etc. Bdmean is the average value of B for all the donor atoms in the first coordination sphere around the metal atom, in this case those within (target distance + 0.5 A).
I am very grateful to Professor Malcolm Walkinshaw, University of Edinburgh, for access to computing facilities, to Dr. Paul Taylor for computational support, and to both for helpful discussions.
Berman,H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. & Bourne,P.E. (2000) Nucleic Acids Research, 28, 235-242.
Bernstein, F.C., Koetzle, T.F., Williams,G.J., Meyer,E.E., Brice,M.D., Rodgers,J.R., Kennard,O., Shimanouchi,T. & Tasumi,M. (1977) J. Mol. Biol. 112, 535.
Degtyarenko,K. (2000) Bioinformatics Review 16, 851-864.
Harding, M.M. (1999) Acta Cryst. D55, 1432-1443.
Harding, M.M. (2000) Acta Cryst. D56, 857-867.
Harding, M.M. (2001) Acta Cryst. D57, 401-411.