Newsletter contents... 
OASIS and Xe phasing: potential in high-throughput crystallography
Quan Hao
Cornell High Energy Synchrotron Source (CHESS), Cornell University,
Ithaca, NY 14853, USA.
Correspondence e-mail: qh22@cornell.edu
The CCP4 supported program OASIS
has been tested using one-wavelength anomalous scattering data collected
from a xenon derivative of the lobster apocrustacyanin A1 protein [Cianci et
al. (2001). The Xe atoms were located by the program SAPI
and the absolute configuration was determined by the program ABS. The electron
density map after OASIS and density
modification clearly revealed the solvent boundary and the C![[alpha]](08_alpha_rmgif.gif) trace. The test demonstrated that, by exploiting the anomalous signal at
single wavelength, OASIS can be used to determine phases at moderate (
trace. The test demonstrated that, by exploiting the anomalous signal at
single wavelength, OASIS can be used to determine phases at moderate (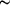 2.3
Å) macromolecular crystallographic resolution for a medium-size protein
(3500 non-H atoms in the asymmetric unit). As the xenon derivatives can
be obtained from native protein crystals using commercially available equipment
in a relatively short time frame (a few hours), the method described in
this paper may provide a good alternative to MAD or MIR phasing, in particular
when high-throughput is desirable. 2.3
Å) macromolecular crystallographic resolution for a medium-size protein
(3500 non-H atoms in the asymmetric unit). As the xenon derivatives can
be obtained from native protein crystals using commercially available equipment
in a relatively short time frame (a few hours), the method described in
this paper may provide a good alternative to MAD or MIR phasing, in particular
when high-throughput is desirable.
Keywords: OASIS; one-wavelength anomalous scattering;
xenon derivative.
|
1. Introduction
In view of the mounting evidence that one-wavelength anomalous scattering
(OAS or otherwise known as SAD) may be sufficient to solve protein structures
(Hao, 2000), the OASIS program (Hao et al, 2000) was written to determine
phases using one-wavelength anomalous scattering data instead of using
additional multiwavelength diffraction data (MAD). This is of particular
importance when protein crystals are sensitive to X-ray irradiation or
the absorption edges of the anomalous scatterers, such as xenon and sulfur,
are difficult to access. A number of minor changes in the new version of
OASIS to be released by CCP4 include new keywords to allow resolution and
sigma cutoff. The upper limit on the number of reflections has been increased
to 150,000 (from 90,000).
In preparing samples for MAD or OAS phasing, the most favored approach
is the incorporation of selenium into protein using seleno-methionine during
the expression of the protein. However, this is only successful when the
gene encoding the particular protein is known and an expression is established
and when the substitution does not affect crystalline order. In cases where
seleno-methionine substitution is not plausible an attractive method for
preparing samples is to incorporate xenon gas into the crystal. Xenon is
known to bind to hydrophobic pockets within proteins at modest pressure.
The one-wavelength anomalous scattering data (courtesy of Dr Rizkallah
and Professor Helliwell, see Table 1 for details) collected from a xenon
derivative of the lobster apocrustacyanin A1 protein (Cianci et
al., 2001) was used to test the possibility of ab initio
phasing.
| Space group |
P212121 |
| Unit cell |
a
= 41.11 Å |
| |
b
= 79.81 Å |
| |
c
= 109.86 Å |
| Non-H atoms in
a.s.u. |
3505 |
| Number of Xe
sites in a.s.u. |
3 major + 1 minor |
| Source |
Daresbury SRS
Station 7.2 |
| Wavelength |
![[lambda]](08_lambda_rmgif.gif) = 2.045 Å
= 2.045 Å |
| f''
(in electrons) |
11.5 |
| Resolution |
64.5-2.3
Å |
| Unique reflections |
16723 |
| Completeness |
99.7% |
| Redundancy |
7.1 |
I/![[sigma]](08_sigma_rmgif.gif) (I) (I) |
23.4 (12.3) |
| Rsym |
7.3 (14.5)% |
|
|
2. Locating the xenon sites
The Se anomalous scatterers for both structures were located by the conventional
direct-methods program
SAPI (Fan
et
al., 1990;
http://staff.chess.cornell.edu/~hao/sapi/sapi.html)
using magnitudes of anomalous differences,
for reflections within 3.0 Å. The solution was selected by
a default run of the program. The largest 416 normalized structure factors
E's
were used in tangent formula phase refinement. The resultant electron density
map produced a group of 3 highest peaks; there was a clear gap between
this group and other peaks in terms of peak height. A Karle-recycle refinement
(an option in SAPI) of these three sites yielded an additional minor site.
The absolute configuration of these sites was determined by the program
ABS (
http://staff.chess.cornell.edu/~hao/abs/abs.html)
based on the
Ps-function
method (Woolfson & Yao, 1994). These Xe sites agreed well with the
published sites (Cianci
et al, 2001) and formed the basis for the
next phasing step.
3. OASIS and DM phasing
The
ab initio phasing of the OAS data
was implemented in the computer program
OASIS
(Hao
et al., 2000). All Friedel pairs
(including centric reflections) were evaluated using
OASIS.
The script that was used to run OASIS is shown below:
-
#oasis.com
-
oasis HKLIN xea1_19_trn.mtz HKLOUT xea1_oasis.mtz << eof
-
TITLE DIRECT PHASING OF xea1 xenon OAS DATA
-
HCO XE 12
-
FIT
-
RES 2.3
-
LCE 7
-
ANO XE 11.5
-
POS XE -0.62360 -0.52335 -0.50331 1 0.318
-
XE -0.87037
-0.55929 -0.99017 2 0.419
-
XE -0.65256
-0.76443 -0.87725 3 0.268
-
XE 0.47078
0.20858 0.15206 4 0.080
-
LABIN F1=F SIGF1=SIGF F2=DANO SIGF2=SIGDANO
-
LABOUT F1=F SIGF1=SIGF PHI=PHIdp W=Wdp
-
END
-
eof
Density modification using the CCP4 program
DM
(Collaborative Computational Project, Number 4, 1994) was then applied
to the resulting phase sets. Phase error analysis and figures of merit
before and after
DM are given in Table
2. The electron density maps after
OASIS
and density modification clearly revealed the solvent boundary. The
C![[alpha]](08_alpha_rmgif.gif)
trace was clearly visible but there were a number of places where the electron
density was broken. A correlation coefficient between the
OASIS
+
DM phased map and the final refined
structure was 0.57.
| |
Phase
errors (°) |
| Number
of reflections |
OASIS |
OASIS
+ DM |
| 3000 |
58.5 |
44.3 |
| 6000 |
59.5 |
47.9 |
| 9000 |
60.0 |
49.6 |
| 12000 |
60.9 |
51.2 |
| 15000 |
61.7 |
52.3 |
| 16723 |
62.1 |
52.9 |
|
|
|
| Mean figure of
merit |
0.49 |
0.71 |
|
|
4. Discussion
Here we demonstrate that, by exploiting the anomalous signal at single
wavelength, OASIS can be used to determine phases at moderate (
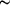
2.3
Å) macromolecular crystallographic resolution for a medium-size protein.
The total CPU time consumed by SAPI, ABS and OASIS was about 3 minutes
on an Alpha XP10000 workstation. As the xenon derivatives can be obtained
from native protein crystals using commercially available equipment in
a relatively short time frame (a few hours), the method described in this
paper may provide an attractive alternative to MAD or MIR phasing, in particular
when high-throughput is desirable.
Acknowledgments
I would like to thank Dr P J Rizkallah and Professor J R Helliwell for
making available the apocrustacyanin A1 data and valuable discussions.
References
Cianci, M., Rizkallah,
P.J., Olczak, A., Raftery, J., Chayen, N.E., Zagalsky., P.F. & Helliwell,
J.R. (2001). Acta Cryst. D57.
1219-1229.
Collaborative Computational Project,
Number 4 (1994). Acta Cryst. D50,
760-763.
Fan, H. F., Hao, Q., Gu, Y. X., Qian,
J. Z., Zheng, C. D. & Ke, H. (1990). Acta Cryst.
A46, 935-939.
Hao, Q., Gu, Y. X., Zheng, C. D. &
Fan, H. F. (2000). J. Appl. Cryst. 33,
980-981.
Woolfson, M. M. & Yao, J. X. (1994). Acta
Cryst. D50, 7-10.
 Newsletter contents...
Newsletter contents... 
![[alpha]](08_alpha_rmgif.gif) trace. The test demonstrated that, by exploiting the anomalous signal at
single wavelength, OASIS can be used to determine phases at moderate (
trace. The test demonstrated that, by exploiting the anomalous signal at
single wavelength, OASIS can be used to determine phases at moderate (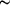 2.3
Å) macromolecular crystallographic resolution for a medium-size protein
(3500 non-H atoms in the asymmetric unit). As the xenon derivatives can
be obtained from native protein crystals using commercially available equipment
in a relatively short time frame (a few hours), the method described in
this paper may provide a good alternative to MAD or MIR phasing, in particular
when high-throughput is desirable.
2.3
Å) macromolecular crystallographic resolution for a medium-size protein
(3500 non-H atoms in the asymmetric unit). As the xenon derivatives can
be obtained from native protein crystals using commercially available equipment
in a relatively short time frame (a few hours), the method described in
this paper may provide a good alternative to MAD or MIR phasing, in particular
when high-throughput is desirable.![[lambda]](08_lambda_rmgif.gif) = 2.045 Å
= 2.045 Å![[sigma]](08_sigma_rmgif.gif) (
(