`fffear' is a package which searches for molecular fragments in poor quality electron density maps. It was inspired by the Uppsala `ESSENS' software (Kleywegt+Jones, 1997), but achieves greater speed and sensitivity through the use of Fast Fourier transforms and a mixed bag of mathematical and computational approaches (Cowtan, 1998). Currently, the main application is the detection of helices in poor electron density maps (5.0A or better), and the detection of beta strands in intermediate electron density maps (4.0A or better).
It is also possible to use electron density as a search model, allowing the location of NCS elements. Approximate matches may be refined, and translation searches may be performed using a single orientation.
The program has also been used to solve phased molecular replacement problems, and to locate missing NCS molecules, although this is not what it was designed for and the results may vary.
The program takes as input an mtz file containing the Fourier coefficients of the map to be searched, and a pdb file (in an arbitrary crystal cell) or map. A `fragment mask' is generated to cover the fragment density, and orientations and translations are searched to find those transformations which give a good fit between the fragment density and map density within the fragment mask.
The program has been highly optimised (Cowtan, 2001) using reciprocal-space rotations and grid-doubling FFT's, and crystallographic symmetry (Rossman+Arnold, 1993) giving 4-50 times speed improvement over the results published in 1998. The speed of the calculation is almost independent of the size of the model, thus the program may also be used for molecular replacement calculations where weak phases are available.
It is also possible to use the program to perform a naive Maximum Likelihood search by using a properly weighted target density and weighted mask. The target density and weighted mask are calculated by a massive search over matching fragments in the PDB. Currently only a 9-resdiue helix is available as a ML target.
The target function is a masked (weighted) mean squared difference between the search density and the current electron density map. As a result, the search function is sensitive to both peaks and voids in the search density. The search density and the map must however be carefully scaled; this is one of the most complex parts of the `fffear' program.
Let the target function be called t(x). The fragment density is pf(x), and the corresponding fragment envelope is ef(x). Then the target function may be formed from the sum of the mean-squared difference in density between the offset fragment and the map: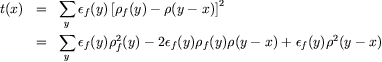
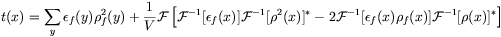
If the Fourier coefficients of the density and squared density are pre-calculated, then the translation function for a fragment in multiple orientations may be calculated by 3 Fast Fourier Transforms (FFTs) per orientation.
The Fourier transform of the search molecule and mask may also be pre-calculated on a fine grid in reciprocal space, and rotated values calculated by interpolation. By this means, the search is reduced to a single FFT per orientation.