Scaling of MAD Data
Philip R. Evans,
MRC Laboratory of Molecular Biology, Hills Road, Cambridge
CB22QH
The integrated intensities from any data collection experiment are not all on
the same scale, because of various systematic differences in the collection
procedure. It is the task of the "data reduction" protocol to place all
observations on a common scale, to detect and reject outliers (reflections for
which the data collection has gone badly wrong), and to produce a list of |F|
and  (|F|) for the structure determination. There are some special
considerations in the optimum treatment of data intended for MAD phasing, in
that we want very accurate differences between amplitudes, for the
anomalous differences
(|F|) for the structure determination. There are some special
considerations in the optimum treatment of data intended for MAD phasing, in
that we want very accurate differences between amplitudes, for the
anomalous differences  F+/- and the dispersive differences
F+/- and the dispersive differences
 F
F , rather than the most
accurate absolute values. This means
a difference both in data collection strategy, designing the experiment to
minimize the systematic errors in the differences, and in the scaling strategy,
in which relative scaling can reduce, though probably not eliminate, the
systematic errors. In the MAD phasing method, we need accurate differences
because the small signal is easily swamped by systematic errors, and we also
need to be careful about eliminating outliers, since a small number of spurious
large differences can confuse both Patterson and direct methods of locating the
anomalous diffracting centres.
, rather than the most
accurate absolute values. This means
a difference both in data collection strategy, designing the experiment to
minimize the systematic errors in the differences, and in the scaling strategy,
in which relative scaling can reduce, though probably not eliminate, the
systematic errors. In the MAD phasing method, we need accurate differences
because the small signal is easily swamped by systematic errors, and we also
need to be careful about eliminating outliers, since a small number of spurious
large differences can confuse both Patterson and direct methods of locating the
anomalous diffracting centres.
To aid designing data collection and scaling strategies, it is helpful to
enumerate the reasons for the observed intensities not being on the same scale.
These factors can be roughly divided into those that can be in principle
calculated, and those that must be determined empirically from the data.
(1) Calculable scale factors
* Lorentz factor - this is uncertain close to the rotation axis, but is not
normally a problem
* Polarization - this may be uncertain for synchrotron radiation, but the error
is small
* Corrections arising from deficiencies in the integration program - if the
geometrical parameters used by the integration program are inaccurate, the
prediction of which spots are partially recorded will also be inaccurate. The
estimated partiality may be improved by post-refinement (eg in Scalepack or
Mosflm)
* Different truncation of the tails of reflections caused by diffuse scattering
- partially recorded reflections are measured over at least twice the rotation
range of fully recorded reflections, so if the spots have long tails in the
rotation direction, more of the tails will be included in partials than in
fulls (the TAILS correction in Scala is an attempt to correct for this, see
appendix below).
(2) Empirical scale factors
These are usually subsumed into general scaling.
* Change of incident beam intensity - mainly on synchrotrons
* Change of detector sensitivity - the variation of sensitivity across the
detector is best determined in a separate calibration (flood-field correction),
but the overall "sensitivity" may be taken up in the scaling, particularly for
film or off-line image plates
* Different crystals
* Illuminated volume - if the crystal is larger than the beam. This is
indistinguishable from absorption in the incident beam
* Absorption - less of a problem at short wavelengths, but hard to correct for
satisfactorily
* Radiation damage - serious on unfrozen crystals
* Wavelength-dependent factors - mainly for the Laue method
It is possible to design the data collection strategy for MAD data collection
such that many of these systematic errors can be made equal, so that they
cancel out in the dispersive and anomalous differences. Note that this is the
opposite of the optimum collection strategy for data intended for structure
refinement, when ideally we should try to maximize the systematic differences
between observations, so that the scaling procedure can determine the different
corrections for different parts of the data, or least can average out the
systematic errors.
(a) Dispersive differences (ie between different wavelengths) - measurements
of the same reflection at different wavelengths will normally be made in the
same way, so that the systematic arrors should be the same. The main difference
is that they are necessarily measured at different times: radiation damage is
the only difficult time-dependent scale, hence the great advantage of using
frozen crystals. On unfrozen crystals, the strategy must be to collect
different wavelengths close together in time (eg as images interleaved at each
wavelength).
(b) Anomalous differences - it is not possible to collect I+ and I- in exactly
the same way, on the same area of the detector. The most difficult correction
is absorption, other corrections are likely to be the same. Absorption is a
serious problem at the longer-wavelength edges (eg Fe), less of a problem for
Se or Br edges.
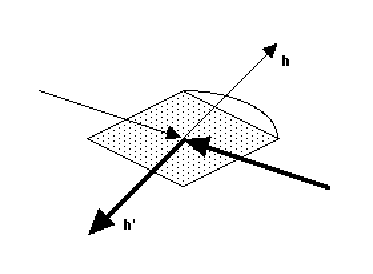
There are two ways of minimizing the absorption differences, though neither
will eliminate the problem:-
(i) inverse beam method - measure reflections at 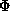 and
and 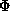 +180deg..
This inverts the direction of the incident and diffracted beams. The absorption
will only be the same if the crystal and its mount have a centre of symmetry
+180deg..
This inverts the direction of the incident and diffracted beams. The absorption
will only be the same if the crystal and its mount have a centre of symmetry
(i)
(ii)
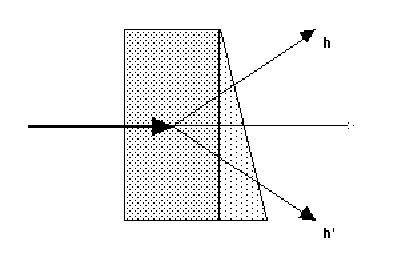
(ii) rotate the crystal about a two-fold axis, and collect Bijvoet pairs (eg
hkl, hk-l for a crystal rotating around the c axis) on the same image. This
requires the crystal to be aligned about an axis, at least approximately. The
absorption is only the same if the crystal and its mount have a plane of
symmetry perpendicular to the rotation axis.
To correct for absorption differences between Bijvoet pairs, the scaling model
must be able to apply a different scale to I+ and to I-, so the scaling model
must be anisotropic and non-centrosymmetric. Suitable functions are
3-dimensional smoothed scales (local scales) and 3D functions such as spherical
harmonics. The functions must not vary too much locally, otherwise the real
differences will be scaled out.
How well are scale factors determined?
The problem with 3-dimensional scale functions is that they are typically
ill-determined by the observed data. The empirical correction factors listed
above may be divided into two categories:-
1) functions of the incident beam direction (illuminated volume,
absorption in the incident beam) or of time,which is equivalent (beam
intensity, radiation damage). With any area detector, these functions are
well-determined, since many reflections are measured at the same time for each
direction.
2) functions of the diffracted beam direction (absorption, radial
dependence of radiation damage). These functions are poorly determined, since
there are relatively few observations in each direction. The corrections are
well-determined only:-
(a) with high symmetry (thus high redundancy of measurements made under
different conditions)
(b) collection by rotation about more than one axis (to measure equivalent
reflections with different beam paths in the crystals)
(c) scaling relative to a reference set - this gives relative rather than
absolute scales, but is useful to reduce systematic errors in differences, as
is required for MAD data.
A relative-scaling protocol
The following suggested protocol for scaling MAD data uses a reference data
set, which provides a an anchor for the scaling parameters. Note that in the
reference, I+ and I- are averaged, so that in the real datasets, systematic
bias in the anomalous difference will be reduced (the mean  I+/- should
be zero). A similar protocol is also useful for scaling heavy-atom derivatives
using the native dataset as reference, in the MIR method.
I+/- should
be zero). A similar protocol is also useful for scaling heavy-atom derivatives
using the native dataset as reference, in the MIR method.
1. Choose reference set: this should be (in order of importance)
(a) the most complete
(b) the most accurate
(c) remote from the anomalous edge
2. Scale and merge the reference set, merging I+ and I-, to get a unique set of
merged intensities Iref
3. Sort the reference set together with all unmerged data, for all wavelengths
(including the set used as reference, if this is to be used in phasing).
4. scale all data together, perhaps in two passes
(a) batch scaling (scale k & B-factor for each image ("batch")) to remove
discontinuities between images. If all images are reliably on a similar scale
with no discontinuities between images (stable source, collected by dose etc),
this step may be omitted.
(b) smooth scaling using a 3-dimensional anisotropic or local scaling model.
This may be parameterized in camera space (x,y,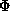 or beam directions) or
in crystal space (h,k,l). An example in Scala would be SCALES ROTATION SPACING
10 DETECTOR 3 3.
or beam directions) or
in crystal space (h,k,l). An example in Scala would be SCALES ROTATION SPACING
10 DETECTOR 3 3.
5. split out each wavelength, either averaging repeated and symmetry-related
observations, or keeping them separate (depending on whether the phasing
strategy uses merged or unmerged data)
Various programs allow scaling of this type, eg the XDS package (Kabsch
1988)), the CCP4 program SCALA (which took some inspiration from the Kabsch
method), and X-GEN (Howard)
Results
Trials with a Se-methionine data set (thanks to Richard Pauptit) and a
Br-uridine DNA set (thanks to Harry Powell and Christine Cardin) showed a small
but significant improvement using this protocol, compared to scaling each
dataset separately. The improvement is presumably only small because
absorption, which causes the most serious systematic errors is small at the Se
and Br edges. Absorption is much more serious at longer wavelength, so for MAD
measurements on for example the Fe edge this scaling method would produce a
much larger gain. However, since the MAD signal is so small, even a small
improvement can make the difference between success and failure, and a small
reduction in the difference between observations (as measured by reduced
dispesive and anomalous differences) may make a substantial difference in
phasing.
Appendix
A simple correction for the bias between fully-recorded and
partially-recorded reflections caused by diffuse scattering
Many protein crystals show marked diffuse scattering, which is seen as long
tails on spots in the "phi" direction, so that reflections often appear on the
image before they are predicted. If the mosaicity is increased to include these
tails, too many reflections may be rejected as overlaps. Fully-recorded
reflections are integrated over a smaller phi width than partials, so more of
the tails are chopped off for fulls than for partials. This leads to the
typical negative partial bias, with partials systematically larger than
equivalent fulls.
A correction has been introduced into SCALA which attempts to correct for the
different truncation of diffuse scattering tails, using a simple model of
thermal diffuse scattering, expressed as 2 or 3 parameters over the whole data
set. This implementation does not attempt to correct for diffuse scattering
itself, only for the different effect on fulls and partials. This correction
reduces the partial bias substantially, and seems to improve the data
generally, though sometimes the parameter refinement can be a little unstable.
The method
This algorithm was inspired by the correction described by Blessing (1987), but
in his case full profiles of the diffraction spots were analysed to determine
the diffuse scattering contribution. Data collected with relatively coarse
rotation slices do not provide enough information to do this, and the typical
crowded diffraction patterns of macromolecule crystals make it harder to
extract full profiles, since the spots may overlap.
1. The thermal diffuse scattering contribution to the integrated intensity is
proportional to the Bragg intensity J. If the complete profile is measured, the
measured intensity I, including diffuse scatter is given by
I = J (1+ )
)
where  is a proportionality constant
is a proportionality constant
2. The proportionality constant  varies with resolution, and may be
anisotropic. At present an isotropic model is implemented in Scala
varies with resolution, and may be
anisotropic. At present an isotropic model is implemented in Scala
 =
=  0 + s2
0 + s2
 1
1
where s2 = (sin 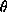 /
/  )2,
and
)2,
and  0
is normally = 0.
0
is normally = 0.  0 and
0 and  1
are refinable parameters.
1
are refinable parameters.
3. The width of the thermal diffuse scattering peak is assumed to be constant
in reciprocal space, = v, a refinable parameter. The distance in reciprocal
space travelled by a reflection rotated by an angle 
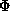 at a
radius
at a
radius 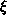 from the rotation axis is given by
from the rotation axis is given by
q = 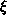

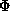
4. The profile of the diffuse scattering peak is modelled as a triangle, with
width v (in the reciprocal space coordinate q), and height h, where h is a
function of  , since the area of the triangle is I-J = h v = J
, since the area of the triangle is I-J = h v = J
 , hence h = J
, hence h = J  / v
/ v
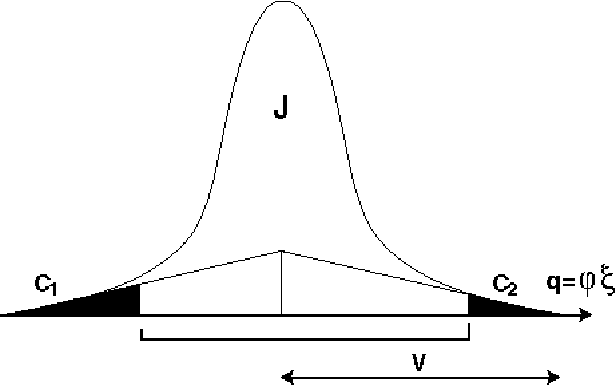
5. If the scan width of an observation, including all parts of a partially
recorded reflection, is less than 2v, the tails of the diffuse scattering peak
may be truncated, clipping off areas C1 and C2 (>= 0) (see figure). These
areas may be calculated from the rotation angles at the start of the scan (the
beginning of the first image contributing to the observation), the centre of
the reflection (the predicted angle), and the end of the scan (the end of the
last image contributing to the observation).
6. The correction factor for diffuse scattering if the full profile were
measured would be given by
J = I / (1 +  )
)
For the truncated profile
J = I / (1 +  ( 1 - C1 - C2))
( 1 - C1 - C2))
where C1 and C2 are expressed as fractions of the complete area of the triangle
(h v)
Since I do not trust this simple formulation to correct properly for diffuse
scattering, the correction used is
J = I (1 +  ) / (1 +
) / (1 +  ( 1 - C1 - C2))
( 1 - C1 - C2))
This corrects for the different truncation of the peak for different spots,
particularly the difference between observations made over 1, 2 ,3 etc images,
but not for the diffuse scattering itself.
The parameters refined are  0,
0,  1 and v (note that C1 and C2
are functions of v), though normally
1 and v (note that C1 and C2
are functions of v), though normally  0 is fixed at 0.0.
0 is fixed at 0.0.
Results
Application of the Tails correction to to datasets with visible diffuse
scattering typically has a dramatic improvement on the partial bias, ie the
systematic difference between fully recorded and partially recorded reflections
(see figure), and often a significant improvement in Rmerge. The correction is
not well-determined if the diffuse scattering is small, nor if the mosaicity is
badly underestimated in the integration process: in these cases, the parameters
can take on unrealistic (eg negative) values.
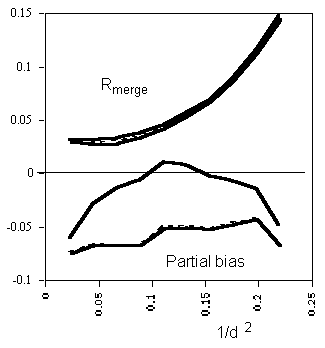
Example of the improvement in partial bias (lower curves) and Rmerge (upper
curves), plotted against resolution. Solid lines: with Tails correction, dashed
lines: without correction. The partial bias is 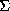 h(<Ifull> -
Ipartial) /
h(<Ifull> -
Ipartial) / 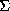 h <Ifull>, where the summations are over all
reflections for which there are both fulls and partials, <Ifull> is the
mean of all the fully recorded observations of the reflection, and Ipartial is
a summed partial observation
h <Ifull>, where the summations are over all
reflections for which there are both fulls and partials, <Ifull> is the
mean of all the fully recorded observations of the reflection, and Ipartial is
a summed partial observation
Acknowledgements: I thank Richard Pauptit, Harry Powell and Christine
Cardin for the loan of datasets, Gérard Bricogne for helpful discussions
on statistics, and Eric de la Fortelle for help in running SHARP.
References
Blessing, R.H. (1987), Data Reduction and Error analysis for Accurate Single
Crystal Diffraction Intensities, Cryst. Rev. 1, 3-58
Howard, A.J. X-GEN documentation
Kabsch, W. (1988) Evaluation of single-crystal X-ray diffraction data from a
position sensitive detector, J. Appl. Cryst. 21, 916-924
 (|F|) for the structure determination. There are some special
considerations in the optimum treatment of data intended for MAD phasing, in
that we want very accurate differences between amplitudes, for the
anomalous differences
(|F|) for the structure determination. There are some special
considerations in the optimum treatment of data intended for MAD phasing, in
that we want very accurate differences between amplitudes, for the
anomalous differences  F+/- and the dispersive differences
F+/- and the dispersive differences
 F
F , rather than the most
accurate absolute values. This means
a difference both in data collection strategy, designing the experiment to
minimize the systematic errors in the differences, and in the scaling strategy,
in which relative scaling can reduce, though probably not eliminate, the
systematic errors. In the MAD phasing method, we need accurate differences
because the small signal is easily swamped by systematic errors, and we also
need to be careful about eliminating outliers, since a small number of spurious
large differences can confuse both Patterson and direct methods of locating the
anomalous diffracting centres.
, rather than the most
accurate absolute values. This means
a difference both in data collection strategy, designing the experiment to
minimize the systematic errors in the differences, and in the scaling strategy,
in which relative scaling can reduce, though probably not eliminate, the
systematic errors. In the MAD phasing method, we need accurate differences
because the small signal is easily swamped by systematic errors, and we also
need to be careful about eliminating outliers, since a small number of spurious
large differences can confuse both Patterson and direct methods of locating the
anomalous diffracting centres. 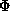 and
and 

 )
)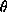 /
/ 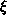 from the rotation axis is given by
from the rotation axis is given by 

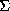 h(<Ifull> -
Ipartial) /
h(<Ifull> -
Ipartial) /